![]() Tiago C. Faria1
Tiago C. Faria1 ![]() ,
, ![]() Claudio Oliveira1,
Claudio Oliveira1, ![]() Iann Leonardo Pinheiro Monteiro2 and
Iann Leonardo Pinheiro Monteiro2 and ![]() Flávio César Thadeo de Lima3
Flávio César Thadeo de Lima3
PDF: EN XML: EN | Supplementary: S1 S2 | Cite this article
Associate Editor: ![]() Carlos DoNascimiento
Carlos DoNascimiento
Section Editor: ![]() William Crampton
William Crampton
Editor-in-chief: ![]() Carla Pavanelli
Carla Pavanelli
Abstract
Uma nova espécie de Hyphessobrycon da bacia do rio Mamuru, Estado do Pará, é descrita. A nova espécie é similar a dois congêneres, Hyphessobrycon hildae e Hy. taguae, e ao não-congênere Hemigrammus bellottii pela presença de uma mancha umeral conspícua e verticalmente alongada, e pela ausência de outras estruturas de coloração escura conspícuas no corpo e nadadeiras, e pela presença, em vida, de um padrão bicolor longitudinal, composto por uma faixa vermelha dorsal e uma faixa iridescente ventral, posicionado dorsalmente a à faixa escura longitudinal. A nova espécie difere de todas estas espécies pela forma da mancha umeral. A metodologia DNA barcoding também distingue a nova espécie das populações amostradas de He. bellottii com distâncias genéticas de 14,97 a 16,8% no gene citocromo c oxidase I, e indica também que a espécie é parte da subfamília Hyphessobryconinae e é espécie-irmã de Hyphessobrycon montagi com distância genética de 9,33%. Comentários sobre o status taxonômico de Hy. hildae e Hy. taguae são apresentados.
Palavras-chave: DNA barcoding, Hemigrammus bellottii, Hyphessobrycon hildae, Hyphessobrycon taguae, Hyphessobryconinae.
Introduction
The genus Hyphessobrycon Durbin, 1908, is the most species rich in Acestrorhamphidae, with 146 species currently recognized as valid (Melo et al., 2024; Toledo-Piza et al., 2024; Lima et al., 2025a,b). The high number of species is related with the high number of lineages included in the genus, which is not only polyphyletic (Mirande, 2019; Ohara et al., 2019; Melo et al., 2024) but also is diagnosed with characters widespread among small acestrorhamphids, i.e., incompletely pored lateral line, caudal fin with scales restricted to its base, two teeth rows on premaxilla with five teeth in the inner row and presence of an adipose fin.
Due to the historic lack of comprehension about the phylogenetic relations of species in Hyphessobrycon, different authors have proposed groups intended as monophyletic within the genus using different combinations of characters, mostly related to color patterns and/or sexual dimorphic characters (e.g., Géry, 1977; Weitzman, Palmer, 1997; Ingenito et al., 2013; Ota et al., 2020). Two of these putative monophyletic groups, the Hyphessobrycon heterorhabdus species-group (Lima et al., 2014; Moreira, Lima, 2017; Faria et al., 2020, 2021) and the Hy. agulha species-group (Géry, 1977; Ohara, Lima, 2015; Faria et al., 2020) are especially relevant in the present context due to their color pattern similarities with the new species described here.
The Hyphessobrycon heterorhabdus species-group is composed, in its most recent proposition (Faria et al., 2021), by Hy. amapaensis Zarske & Géry, 1998, Hy. cantoi Faria, Guimarães, Rodrigues, Oliveira & Lima, 2021, Hy. ericae Moreira & Lima, 2017, Hy. heterorhabdus (Ulrey, 1894), Hy. montagi Lima, Coutinho & Wosiacki, 2014, Hy. sateremawe Faria, Bastos, Zuanon & Lima, 2020, and Hy. wosiackii Moreira & Lima, 2017. This species-group is diagnosed by the presence of a tricolor longitudinal pattern composed by: a dorsal red stripe, a middle iridescent stripe and a ventral dark longitudinal pattern composed by a black humeral blotch, which is well-defined anteriorly and diffuse posteriorly, followed posteriorly by a black stripe that becomes blurred towards the caudal peduncle (the latter absent in most Hy. ericae populations and in Hy. wosiackii; see Moreira, Lima, 2017). In its turn, the Hy. agulha species-group is characterized by the presence of a dark, broad, and diffuse longitudinal stripe that occupies most of the ventral half of the body. Currently, the following species are considered to belong to this group: Hy. agulha Fowler, 1913, Hy. clavatus Zarske, 2015, Hy. eschwartzae García-Alzate, Román-Valencia & Ortega, 2013, Hy. herbertaxelrodi Géry, 1961, Hy. klausanni Garcia-Alzate, Urbano-Bonilla & Taphorn, 2017, Hy. loretoensis Ladiges, 1938, Hy. lucenorum Ohara & Lima, 2015, Hy. margitae Zarske, 2016, Hy. metae Eigenmann & Henn, 1914, Hy. mutabilis Costa & Géry, 1994, Hy. peruvianus Ladiges, 1938, Hy. wadai Marinho, Dagosta, Camelier & Oyakawa, 2016 and Hy. zoe Faria, Lima & Wosiacki, 2020 (Faria et al., 2020).
However, the recent broad phylogenetic analysis of the former family Characidae presented by Melo et al. (2024) has not recovered either the Hy. heterorhabdus (sensu Faria et al., 2021) nor the Hy. agulha (sensu Faria et al., 2020) as monophyletic. Melo et al. (2024:19, fig. 6) analyzed three species assigned to the Hy. heterorhabdus species group (sensu Faria et al., 2021), i.e., Hy. amapaensis, Hy. ericae, and Hy. heterorhabdus, and six species belonging to the Hy. agulha species group (sensu Faria et al., 2020), i.e., Hy. agulha, Hy. herbertaxelrodi, Hy. margitae, Hy. mutabilis, Hy. peruvianus, and Hy. wadai. Species of both the Hy. heterorhabdus species group and the Hy. agulha species-group were recovered intermigled in a clade within the newly proposed subfamily Hyphessobryconinae that also included Hy. rubrostriatus and Hemigrammus bellottii, and in addition several unidentified Hyphessobrycon species. A single species, Hy. wadai, was recovered outside this clade, within its sister-clade, as the sister-species of Hy. cyanotaenia. In the present work, we describe a new species of Hyphessobrycon,which is part of subfamily Hyphessobryconinae (sensu Melo et al., 2024), specifically of the clade that includes species belonging to the Hy. heterorhabdus and Hy. agulha species groups. We use DNA barcoding methodology to investigate its phylogenetic relations with the most similar species of the clade, including different populations of He. bellottii, a similar-looking syntopic species, and Hy. montagi, a species suggested to be related to the Hy. heterorhabdus species group on its original description (Lima et al., 2014). The new taxon is also the first species described from the rio Mamuru basin, a small tributary of the Amazon basin draining an area immediately west to the rio Tapajós basin and discharging into the Paraná de Ramos channel, a ria lake situated in the intervening area between the lower rio Tapajós and the lower rio Madeira.
Material and methods
Counts and measurements follow Fink, Weitzman (1974), except for the number of horizontal scale rows below lateral line, which are counted to the pelvic-fin insertion (excluding the axillary scale) rather than to the anal-fin origin, and the addition of three measurements: distance from pelvic-fin origin to anal-fin origin, dorsal-fin base length, and anal-fin base length. Standard length (SL) is expressed in millimeters (mm) and all other measurements are expressed as percentages of SL, except subunits of the head, which are expressed as percentages of head length (HL). In the description, counts are followed by their absolute frequency in parentheses. Asterisks indicate the counts of the holotype. Circulii and radii were counted on scales from the row immediately dorsal to the lateral line at the vertical through the dorsal-fin origin. Counts of supraneurals, branchiostegal rays, gill-rakers of the first branchial arch, teeth counts (with exception of premaxillary teeth) and morphology, unbranched anal-fin rays, procurrent caudal-fin rays and position of pterygiophores were taken from cleared and stained (c&s) specimens prepared according to Taylor, Van Dyke (1985). Vertebrae of the Weberian apparatus were counted as four elements and the compound caudal centrum (PU1+U1) as a single element. Catalog numbers are followed by the total number of specimens and their SL range. The number of c&s specimens is given in parentheses, followed by their respective SL range. Institutional abbreviations follow Sabaj (2020, 2023).
Molecular analysis. DNA extraction followed Ivanova et al. (2006) and partial sequences of the mitochondrial gene cytochrome c oxidase subunit I (COI) were amplified by polymerase chain reaction (PCR), with primers FishF1/R1 described by Ward et al. (2005). Reactions were carried out in a 12.5 μL reaction volume containing 1.25 μL of 10× PCR buffer, 0.40 μL MgCl2 (50 mM), 0.30 μL dNTPs (2 mM), 0.25 μL of each primer (5 μM), 0.20 μL of PHT Taq DNA polymerase (Phoneutria), and 2 μL DNA template (200 ng), and 7.85 μL of ddH2O. The PCR consisted of denaturation (5 min at 95°C) followed by 30 cycles of denaturation (1 min at 95°C), primer hybridization (45 sec at 52°C), nucleotide extension (1 min at 68°C), and a final extension (10 min at 68°C). All PCR products were checked using 1% agarose gel and purified with ExoSap-IT (USB Corporation) following the manufacturer’s instructions. The purified PCR products were sequenced using the Big DyeTM Terminator v. 3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Austins, USA), and purified through ethanol precipitation. Amplified fragments were then loaded into an ABI 3500 Genetic Analyzer (Applied Biosystems), in the Instituto de Biotecnologia (IBTEC), Instituto de Biociências, Universidade Estadual Paulista “Júlio de Mesquita Filho”, Botucatu, Brazil. For this study, we generated two sequences of the new species and 13 sequences of other nine nominal species of Hyphessobrycon and Hemigrammus. We also used two sequences of Hy. heterorhabdus obtained from Genbank. For more details about sequences and Genbank numbers, see Tab. 1. The sequences were assembled using the software Geneious v. 7.1.4 (Kearse et al., 2012) and aligned with Muscle (Edgar, 2004) under default parameters. The best-fit model of nucleotide evolution was selected according to Akaike Information Criterion with corrections for small sample sizes (AICc). The overall mean genetic distances (among all specimens), as well as interspecific (among species group) and intraspecific distances (among specimens of each species group), were estimated with 1.000 pseudoreplicates and without root. These previous analyses were estimated using MEGA v. 11. Maximum likelihood (ML) analysis was performed in RAxML-HPC v. 8 on ACCESSusing the GTRGAMMA model in the CIPRES server. The best tree was accessed through ten random searches with 1,000 bootstrap pseudoreplicates. The resulting ML tree was used as an input tree for the Poisson Tree Process model (PTP) analysis (Zhang et al., 2013), which was performed on the PTP web server (https://species.h-its.org), with the option “remove outgroup” and the others parameters in default. The analysis of Assemble Species by Automatic Partitioning (ASAP) (ASAP; Puillandre et al., 2021) is available in the ASAP webserver (https://bioinfo.mnhn.fr/abi/public/asap/asapweb.html) with model Jukes-Cantor (JC69).
TABLE 1 | Voucher information for analyzed sequences, and Genbank or BOLD numbers.
Species | Collection | Voucher | River basin | Specific location | Municipality/ State | Coordinates | BOLD | Genbank |
Hemigrammus bellottii | LBP 33007 | 112819 | Rio Abacaxis | Rio Abacaxis | Maués, Amazonas |
06°41’51.58”S |
| PP786517 |
Hemigrammus bellottii | LBP 33193 | 110911 | Rio Mamuru | Rio Mamuru | Itaituba, Pará |
03°59’51.13”S |
| PP786518 |
Hemigrammus bellottii | LBP 33193 | 110910 | Rio Mamuru | Rio Mamuru | Itaituba, Pará |
03°59’51.13”S |
| PP786519 |
Hemigrammus bellottii | LBP 22476 | 86809 | Rio Amazonas | Quebrada La Ponderosa | Leticia, Colombia |
04°08’24.4”S |
| PQ044052 |
Hyphessobrycon agulha | LBP 30463 | 50120 | Rio Madeira | Igarapé da UNIR | Porto Velho, Rondônia |
08°49’36.1”S |
| PP786516 |
Hyphessobrycon amapaensis | LBP 30508 | 116901 | Rio Amazonas | Igarapé affluent of rio Pedreira | Ferreira Gomes, Amapá |
00°41’44.0”N |
| PP372830 |
Hyphessobrycon cf. ericae | LBP 31628 | 110207 | Rio Curuá-Una | Rio Moju | Belterra, Pará |
03°25’05.6”S |
| PP372834 |
Hyphessobrycon cf. ericae | LBP 31628 | 110208 | Rio Curuá-Una | Rio Moju | Belterra, Pará |
03°25’05.6”S |
| PP372835 |
Hyphessobrycon eschwartzae | ROMCID 134066 | 10194 | Rio Madre de Dios | Rio Planchon | Tambopata, Madre de Dios, Peru* |
12°16’37.67”S |
| PQ044050 |
Hyphessobrycon herbertaxelrodi | LBP 8387 | 40453 | Rio Paraguai | Córrego Águas Claras | Tangará da Serra, Mato Grosso |
14°21’03.2”S |
| PP372838 |
Hyphessobrycon heterorhabdus | LBP 9451 | 45139 | Rio Guamá | Igarapé affluent of igarapé São José | Santa Isabel, Pará |
01°16’54.1”S | ACD9730 |
|
Hyphessobrycon heterorhabdus | LBP 9451 | 45140 | Rio Guamá | Igarapé affluent of igarapé São José | Santa Isabel, Pará |
01°16’54.1”S | ACD9730 |
|
Hyphessobrycon montagi | LBP 31642 | 110260 | Rio Arapiuns, Tapajós | Aquarium specimen |
|
|
| PP786520 |
Hyphessobrycon mamuruensis | LBP 33015 | 112842 | Rio Mamuru | Igarapé affluent of rio Mamuru | Itaituba, Pará |
04°04’46.03”S |
| PP786521 |
Hyphessobrycon mamuruensis | LBP 33015 | 112841 | Rio Mamuru | Igarapé affluent of rio Mamuru | Itaituba, Pará |
04°04’46.03”S |
| PP786522 |
Hyphessobrycon pulchripinnis | LBP 32962 | 112719 | Rio Tapajós | Igarapé do Abacaxi | Jacareacanga, Pará |
06°08’51.9”S |
| PP372832 |
Hyphessobrycon sateremawe | INPA 50729 | 30449 | Rio Abacaxis | Igarapé affluent of rio Abacaxis | Nova Olinda do Norte, Amazonas |
04°17’5”S |
| PQ044051 |
Results
Hyphessobrycon mamuruensis, new species
urn:lsid:zoobank.org:act:1A4F8098-AD6B-4DC6-AB26-E92A443D35A8
(Figs. 1–2; Tab. 2)
Holotype. LBP 34880, 32.4 mm SL, Brazil, Pará State, Itaituba, igarapé tributary of rio Mamuru, near comunidade Mamuru, 04º02’19.66”S 56º15’24.5”W, T. C. Faria & I. L. P. Monteiro, 12 Dec 2022.
Paratypes. All from Brazil, Pará State, Itaituba. LBP 33134, 53, 16.5–32.0 mm SL, MPEG 39787, 5, 25.2–29.1 mm SL, same data as holotype. LBP 33024, 148, 15.4–32.8 mm SL, ZUEC 17660, 10, 22.4–31.2 mm SL, INPA 61051, 10, 23.7–30.6 mm SL, MZUSP 130842, 5, 19.8–30.9 mm SL, Igarapé tributary of rio Mamuru, 04º04’46”S 56º11’45.41”W, T. C. Faria & I. L. P. Monteiro, 12 Dec 2022. LBP 33188, 27, 18.6–26.8 mm SL, rio Mamuru, 03º59’51”S 56º16’54.5”W, T. C. Faria & I. L. P. Monteiro, 2 Dec 2022. LBP 33177, 23, 21.0–30.4 mm SL, Igarapé tributary of rio Mamuru, 04º03’29.3”S 56º13’24.2”W, T. C. Faria & I. L. P. Monteiro, 12 Dec 2022. LBP 33388, 107, 15.9–35.5 mm SL, 4 c&s, same locality and collectors as holotype, 2 Dec 2022.
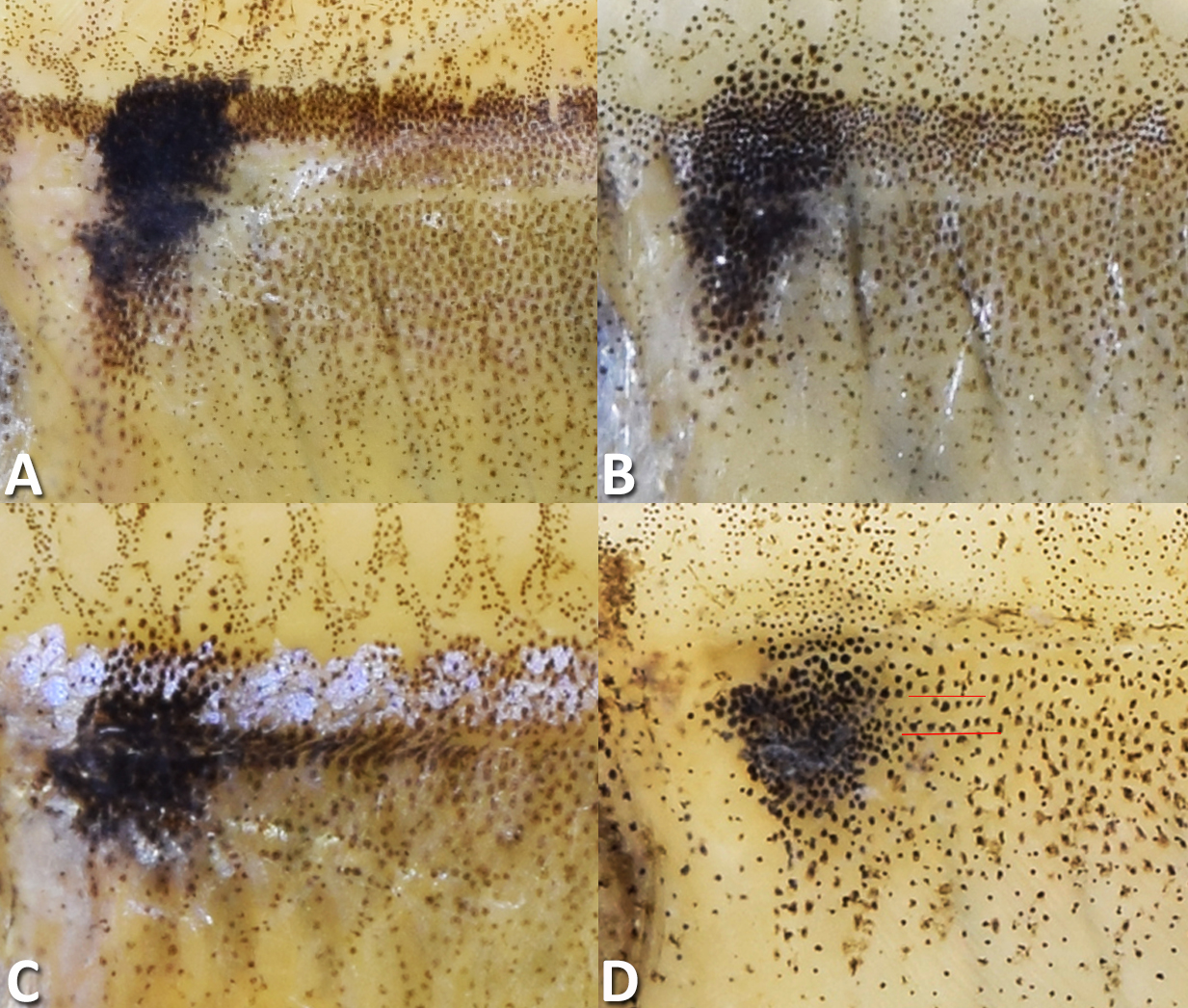
Diagnosis. Hyphessobrycon mamuruensis can be distinguished from all congeners, except Hy. bussingi Ota, Carvalho & Pavanelli, 2020, Hy. columbianus Zarske & Géry, 2002, Hy. condotensis Regan, 1913, Hy. daguae Eigenmann, 1922, Hy. ecuadorensis (Eigenmann, 1915), Hy. eilyos Lima & Moreira, 2013, Hy. hildae Fernandez-Yépez, 1950, Hy. igneus Miquelarena, Menni, López & Casciotta, 1980, Hy. itaparicensis Lima & Costa, 2001, Hy. panamensis Durbin, 1908, Hy. pinnistriatus Carvalho, Cabeceira & Carvalho, 2017, Hy. platyodus Ohara, Abrahão & Espínola, 2017, Hy. pulchripinnis Ahl, 1937, Hy. rheophilus Ohara, Teixeira, Albornoz-Garzón, Mirande & Lima, 2019, and Hy. taguae García-Alzate, Román-Valencia & Taphorn, 2010, by the combination of the following color pattern features: presence of a single humeral blotch, lack of a conspicuous caudal-peduncle blotch, lack of a conspicuous blotch on the dorsal fin and lack of a conspicuous longitudinal dark stripe (vs. presence of two humeral blotches, or absence of any humeral blotch, presence of a conspicuous dark blotch on the caudal peduncle, presence of a conspicuous blotch on the dorsal fin and/or presence of a conspicuous longitudinal dark stripe). Hyphessobrycon mamuruensis can be distinguished from all species belonging to the Hy. panamensis species-group (sensu Ota et al., 2020; i.e., Hy. bussingi, Hy. columbianus, Hy. daguae, Hy. ecuadorensis, and Hy. panamensis) by lacking bony hooks on the anal fin of dimorphic males (vs. presence of individual large bony hooks on each anterior large ray the anal fin). Hyphessobrycon mamuruensis can be additionally distinguished from Hy. bussingi, Hy. columbianus, Hy. condotensis, Hy. daguae, Hy. ecuadorensis, Hy. eilyos, Hy. igneus, Hy. itaparicensis, Hy. panamensis, Hy. pinnistriatus, Hy. platyodus, Hy. pulchripinnis, Hy. rheophilus, by the presence well-defined humeral blotch in preserved specimens (vs. presence of a relatively inconspicuous, diffuse humeral blotch). Hyphessobrycon mamuruensis can additionally be distinguished from Hy. platyodus by presenting the anal fin in mature males with anteriormost anal-fin rays distinctly longer forming an anterior lobe (vs. mature males with anal-fin rays roughly equal in size along its length and lacking any distinct fin lobe). Hyphessobrycon mamuruensis can be distinguished from Hy. hildae, Hy. taguae, and from the non-congener but similar-looking Hemigrammus bellottii, by the presence of a well-developed, vertically elongated humeral blotch considerably higher than wide (vs. humeral blotch only slightly higher than wide in Hy. hildae, Hy. taguae,and He. bellottii; see Fig. 3). It is further distinguished from Hy. taguae and He. bellottii, by the absence of a posterior extension of the humeral blotch (vs. presence of a thin, short posterior extension of the humeral blotch; see Fig. 3), and by lacking bony hooks on the anterior rays of the anal fin of mature males (vs. presence of bony hooks on the anterior rays of the anal fin of mature males). See the Discussion, for further notes on both Hy. hildae and Hy. taguae.
FIGURE 1| Hyphessobrycon mamuruensis. A. LBP 34880, holotype, 32.4 mm SL, female; B. LBP 33134, paratype, 24.4 mm SL, male, Brazil, Pará, rio Mamuru.
FIGURE 2| Hyphessobrycon mamuruensis. Live specimen, paratype, LBP 33024, SL uncertain, same locality as holotype.
FIGURE 3| Humeral blotches of Hyphessobrycon mamuruensis (A, B) and Hemigrammus bellottiii specimens (C, D). A. LBP 34880, holotype, 32.4 mm SL, showing no posterior extension. B. LBP 33134, paratype, 24.4 mm SL, showing no posterior extension. C. LBP 34342, 37.2 mm SL, showing conspicuous posterior extension. D. LBP 34285, 28.7 mm SL, showing inconspicuous posterior extension (between red lines).
Description. Morphometric data for holotype and paratypes in Tab. 2. Body compressed. Greatest body depth at vertical through dorsal-fin origin. Dorsal profile of the head slightly convex from upper lip to vertical through posterior nostril, straight from that point to tip of supraoccipital spine. Dorsal profile of body slightly convex from latter point to anterior terminus of dorsal fin. Dorsal-fin base straight, posteroventrally slanted, slightly convex from posterior terminus of dorsal fin to adipose-fin insertion and slightly concave between adipose-fin insertion and origin of anteriormost dorsal procurrent caudal-fin ray. Ventral profile of head and body convex from tip of lower jaw to anal-fin origin. Anal-fin base straight, posterodorsally slanted. Ventral profile of caudal peduncle slightly concave.
TABLE 2 | Morphometric data for Hyphessobrycon mamuruensis. N = number of specimens measured; SD = Standard deviation.
| Holotype | Range | Mean±S.D. | N |
Standard length(mm) | 32.4 | 20.0–32.8 | – | – |
Percents of standard length | ||||
Depth at dorsal-fin origin | 34.6 | 27.5–35.0 | 32.1±1.7 | 29 |
Snout to dorsal-fin origin | 49.7 | 48.4–50.6 | 49.7±0.7 | 29 |
Snout to pelvic-fin origin | 47.5 | 44.9–49.3 | 47.2±1.1 | 29 |
Snout to anal-fin origin | 62.3 | 60.5–65.4 | 62.7±1.1 | 29 |
Caudal peduncle depth | 8.9 | 7.0–10.0 | 8.6±0.8 | 29 |
Caudal peduncle length | 14.8 | 10.5–15.2 | 13.3±1.1 | 29 |
Pectoral-fin length | 22.8 | 20.1–24.1 | 22.8±0.7 | 29 |
Pelvic-fin length | 18.2 | 15.2–20.7 | 19.1±1.2 | 29 |
Dorsal-fin base | 13.9 | 12.4–14.7 | 13.8±0.6 | 29 |
Dorsal-fin length | 32.7 | 29.5–33.6 | 32.3±0.9 | 29 |
Anal-fin base | 29.0 | 26.5–29.5 | 28.1±0.8 | 29 |
Head length | 26.8 | 26.5–29.0 | 27.7±0.7 | 29 |
Percents of head length | ||||
Horizontal orbital diameter | 40.2 | 36.9–44.3 | 41.6±1.7 | 29 |
Snout length | 23.0 | 20.0–25.9 | 22.9±1.7 | 29 |
Least interorbital width | 31.0 | 28.4–33.3 | 30.7±1.3 | 29 |
Upper jaw length | 43.7 | 40.5–47.4 | 44.8±1.6 | 29 |
Jaws equal, mouth terminal. Posterior terminus of maxilla reaching vertical through anterior margin of iris. Maxilla approximately at 45 degrees angle relative to longitudinal axis of body. Nostrils close to each other, anterior opening oval, posterior opening crescent-shaped. Premaxillary teeth in two rows. Outer teeth row with 2(2), 3(10), or 4(2) conic to tricuspid teeth. Inner row with 5(13) or 6(1) bi- to pentacuspid teeth, symphyseal tooth narrower than neighbor tooth. Maxilla with 2(2) or 3(2), conical to tricuspid teeth. Dentary with 10(1), 12(1) or 13(2) teeth, anteriormost 3–4 teeth larger, bi- to tricuspid, one teeth uni- to tricuspid intermediary in size, remaining teeth considerably smaller and conical. Central cusp of all teeth more developed than lateral cusps.
Scales cycloid. Two to seven radii strongly marked, circulii well-marked anteriorly, weakly marked posteriorly. Lateral line slightly deflected downward and incompletely pored, with 7(6), 8*(18) or 9(5) perforated scales. Longitudinal scales series including lateral-line scales 32*(10), 33(10), 34(6) or 35(2). Longitudinal scale rows between dorsal-fin origin and lateral line 5*(28) or 6(1). Longitudinal scale rows between lateral line and pelvic-fin origin 3(18) or 4*(11). Predorsal scales 9(7), 10*(18) or 11(4). Circumpeduncular scales 12*(26). Caudal fin with few small scales basally.
Dorsal-fin rays ii, 8*(1) or 9(28). Dorsal-fin origin approximately at middle of standard length. First dorsal-fin pterygiophore inserting behind neural spine of 8th(1) or 9th(3) vertebrae. Adipose fin present. Anteriormost anal-fin pterygiophore inserting posterior to haemal spine of 15th(3) or 16th(1) vertebrae. Anal-fin rays iii(1) or iv(3), 18(3), 19(9), 20*(13), 21(3) or 22(1). Last unbranched and first to third anteriormost branched rays distinctly longer than remaining rays, subsequent rays gradually decreasing in size. Pectoral-fin rays i,9(3), 10*(15), 11(10) or 12(1). Pelvic-fin rays i,7*(29). Tip of pelvic fin reaching anteriormost anal-fin rays. Caudal fin forked, lobes roughly rounded and of similar size. Nine (1), 10(1) or 11(2) dorsal procurrent caudal-fin rays, and 8(3) or 9(1) ventral procurrent caudal-fin rays. Vertebrae 33(4).
Supraneurals 4(3) or 5(1), upper portion wider. Branchiostegal rays 4. First gill arch with 1(1) or 2(3) hypobranchial, 1(1) on cartilage between hypobranchial and ceratobranchial, 7(1), 8(2) or 9(1) ceratobranchial, 1(3) on cartilage between ceratobranchial and epibranchial, and 4(1), 5(2) or 6(1) epibranchial gill-rakers.
Coloration in alcohol. Overall body color beige. Dorsal half of body darker. Dorsal portion of head dark. Ventralmost portion of head and body with low concentration of scattered dark chromatophores. Snout and tip of dentary dark. Numerous dark chromatophores across infraorbitals, maxilla and opercle. Predorsal and preadipose scales with conspicuous central dark blotches. Predorsal-scale peripheral region less darker than its center. Scale rows dorsal to midbody with conspicuous reticulated pattern formed by dark chromatophores concentrated at scales margins. Humeral blotch vertically elongated, roughly triangular, about two scales high and one to one and a half wide, with smallest angle directed downward. Humeral blotch surrounded anteriorly and posteriorly by clear areas lacking melanophores. Narrow, ill-defined longitudinal stripe, conspicuous only in some specimens (e.g., Fig. 1A), extending from upper portion of humeral blotch to vertical through adipose fin insertion, fading considerably after vertical through middle portion of dorsal fin. Relatively dense concentration of dark chromatophores along the midbody, after the humeral blotch, imparting an overall dark coloration and contrasting with the markedly clearer ventral area. Narrow dark line along midlateral septum extending from vertical through middle portion of dorsal fin to anterior portion of caudal peduncle. Dark chromatophores aligned along myocommata of hypaxial muscles above anal fin and parallel to anal fin base. Pectoral fin and pelvic fin hyaline with few scattered dark chromatophores. Dorsal-fin ray hyaline, with dark chromatophores concentrated on anteriormost rays and distal region. Anal fin hyaline, with dark chromatophores scattered along interradial membranes. Adipose fin with few scattered dark chromatophores, mainly at its base. Caudal fin mostly hyaline, with few dark chromatophores scattered on interradial membranes, with a higher concentration on distalmost regions and along ventralmost and dorsalmost region of fin lobes.
Coloration in life. Based on pictures of living specimens taken at the field (Fig. 2): overall body color olivaceous. Lower half of head and abdominal region silvery. Dorsal portion of eye red. Two longitudinal, juxtaposed stripes on flanks, one dorsal red stripe and one ventral iridescent stripe. Stripes extending on midlateral region immediately dorsal to darkened midbody area (see coloration in alcohol). Red stripe, anteriorly thicker and continuous, becoming row of red blotches after dorsal fin insertion, ending in large red blotch at posterodorsal region of caudal peduncle. Iridescent stripe golden, continuous, more diffuse on caudal peduncle, ending immediately before red blotch. Humeral blotch vertically elongated, conspicuous, and with diffuse borders. Humeral blotch dorsal expansion with about half the height of ventral expansion. Iridescent bluish chromatophores immediately ventral to midlateral golden stripe and scattered on dorsalmost portion of abdominal region. Dorsal and adipose fin, anterior lobe of anal fin, proximal half of caudal-fin lobes and anterior rays of pelvic fins yellow.
Sexual dimorphism. Females present the last unbranched and two anteriormost branched anal-fin rays longer, resulting in a more pointed and developed anal-fin lobe. Males present tiny bony hooks restricted to the pelvic fins, along all rays, an uncommon condition among acestrorhamphids recently discussed by Lima et al. (2025b:11). Females reach larger sizes than males (largest female 35.5 mm SL, largest male 26.5 mm SL). The presence of female-biased sexual size dimorphism in characiforms was recently discussed by Teixeira et al. (2025:140).
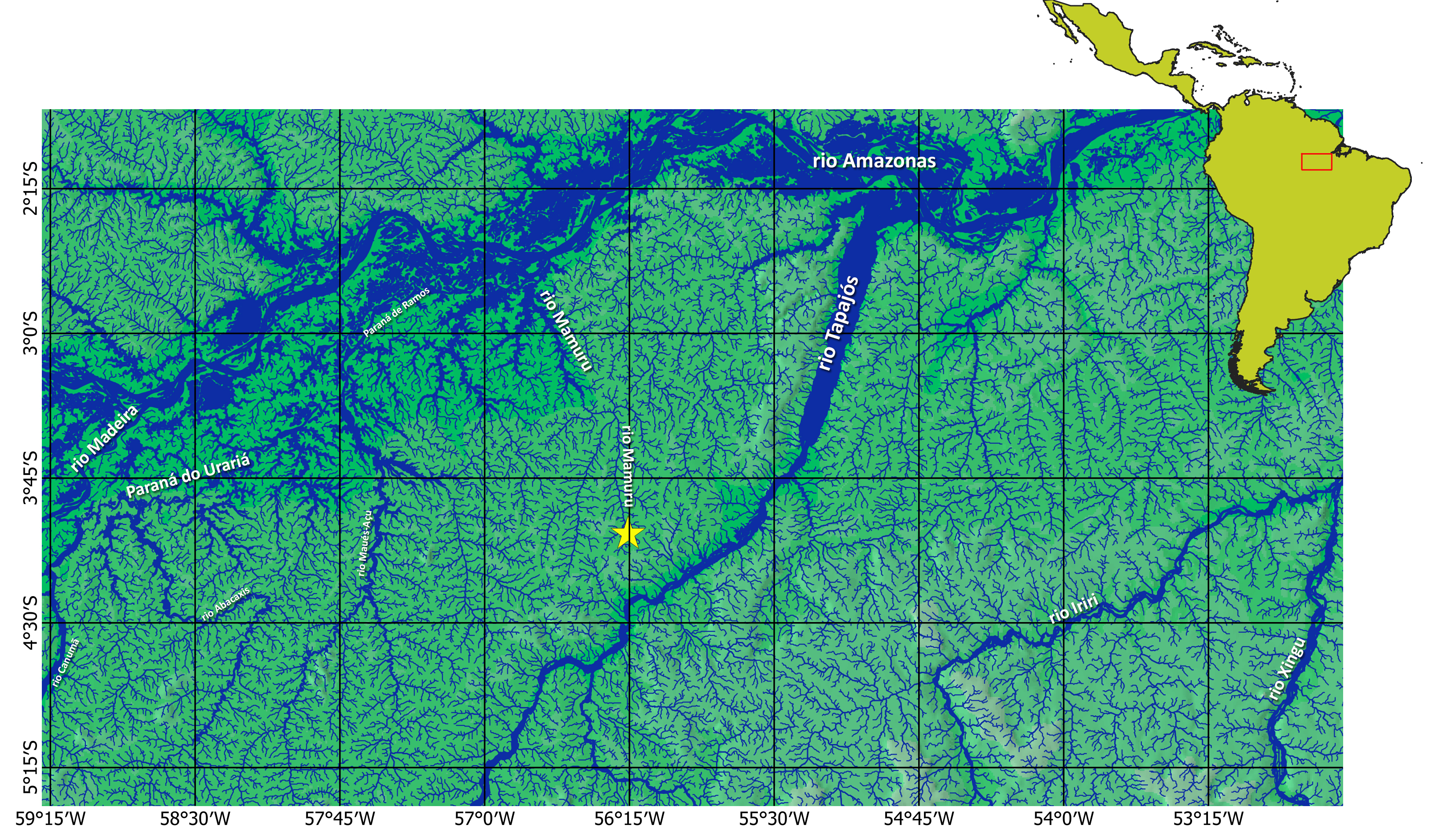
Geographical distribution. Hyphessobrycon mamuruensis is known only from the upper rio Mamuru basin, State of Pará, near the border with Amazonas State, Brazil (Fig. 4).
FIGURE 4| Map of the central Amazon basin, showing the known distribution of Hyphessobrycon mamuruensis (yellow star comprises multiple localities).
Ecological notes. Hyphessobrycon mamuruensis is known from moderate-flowing, clear water forest streams with sandy bottom (Fig. 5). Specimens were collected in both the main channel of upper rio Mamuru and in its small tributaries. It was the most common species in smaller tributaries, where larger specimens were also more abundant. Hemigrammus bellottii, the most similar-looking species to Hy. mamuruensis (see Discussion), was collected syntopically in most localities, but was less abundant than the latter at the rio Mamuru basin.
FIGURE 5| A and B. Type-locality of Hyphessobrycon mamuruensis, a small tributary of rio Mamuru. C. Main channel of rio Mamuru.
Etymology. The specific epithet is a reference to type-locality of the new species, rio Mamuru basin. A noun in apposition.
Conservation status. Hyphessobrycon mamuruensis is known only from the upper rio Mamuru basin. The region of occurrence of the species is right at the frontier of the Amazon’s deforestation arc. In fact, Itaituba is a hotspot in the Amazon for gold mining and deforestation and there are roads connecting the city to the upper portion of rio Mamuru basin. Fortunately, the species’ known range is very close to the Parque Nacional da Amazônia, which protects the left bank tributaries of rio Mamuru basin. Due to this, we suggest the conservation status of the new species as Low Concern (LC) according to the IUCN criteria and categories (2024).
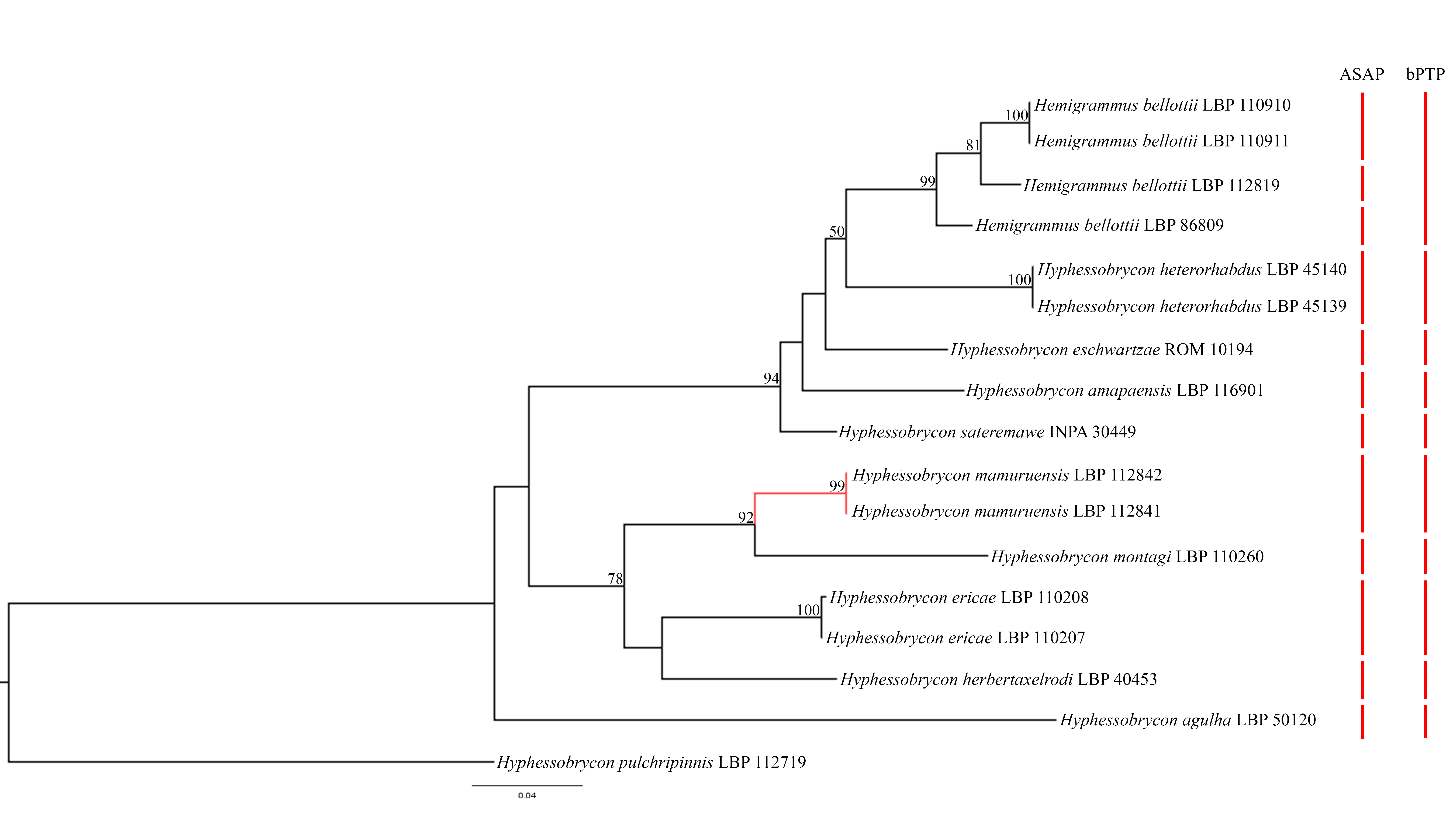
Genetics. The recovered tree using DNA barcoding methology (Fig. 6) indicates Hyphessobrycon mamuruensis and Hemigrammus bellottii as not closely related, with the two belonging to different groups with high bootstrap support. This result contrasts with the fact that He. bellottii is the most similar-looking species to Hy. mamuruensis known to date.
FIGURE 6| RAxML tree based on DNA barcoding methodology showing Hyphessobrycon mamuruensis (red clade) and similar species. Species delimitation methods indicate the presence of 9 or 10 species in the ingroup.
Hemigrammus bellottii is recovered as monophyletic, but ASAP species delimitation method indicates each population sampled (from Tabatinga, its type-locality, rio Mamuru and rio Abacaxis basins) as different species while bPTP indicates all populations as a single species (Figs. S1, S2). All other nominal species are recovered as a single evolutionary lineage by both species delimitation methods. A clade consisting of Hy. mamuruensis, Hy. herbertaxelrodi, Hy. ericae and Hy. montagi is recovered with 78% bootstrap support, indicating a sister relation between Hy. mamuruensis and Hy. montagi with high bootstraps support (92%).
A sister clade relationship between He. bellottii and Hy. heterorhabdus is recovered with small bootstraps support (50%). The Hy. heterorhabdus species group (sensu Lima et al., 2014; Moreira, Lima, 2017; Faria et al., 2020, 2021) is recovered as non-monophyletic with the inclusion of He. bellottii and Hy. eschwartzae and exclusion of Hy. montagi and Hy. ericae. A clade consisting of He. bellottii, Hy. sateremawe, Hy. heterorhabdus, Hy. amapaensis and Hy. eschwartzae is recovered with bootstrap support of 98%.
The genetic distances among the ingroup samples (Tab. 3; not considering genetic distances between same species samples recovered by ASAP) ranged between 20.42% (between Hy. agulha and He. bellottii from rio Mamuru) and 2.97% (between He. bellottii from rio Mamuru and He. bellottii from rio Abacaxis). The genetic distances between He. bellottii and Hy. mamuruensis range between 14.97% and 16.8% (respectively between Hy. mamuruensis and populations of He. bellottii from Tabatinga and rio Abacaxis). The genetic distance between Hy. mamuruensis and Hy. montagi is 9.33%.
TABLE 3 | Genetic distances based on DNA barcoding using Maximum Composite Likelihood among species Hyphessobrycon and Hemigrammus similar to Hyphessobrycon mamuruensis.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
1- Hy. pulchripinnis |
|
|
|
|
|
|
|
|
|
|
|
|
2- Hy. agulha | 24.14% |
|
|
|
|
|
|
|
|
|
|
|
3- Hy. heterorhabdus | 21.56% | 18.97% |
|
|
|
|
|
|
|
|
|
|
4- Hy. amapaensis | 24.52% | 18.74% | 11.00% |
|
|
|
|
|
|
|
|
|
5- Hy. eschwartzae | 22.66% | 18.57% | 9.26% | 8.73% |
|
|
|
|
|
|
|
|
6- Hy. sateremawe | 22.50% | 18.78% | 8.77% | 7.39% | 7.13% |
|
|
|
|
|
|
|
7- He. bellottii Leticia (type-locality) | 22.85% | 19.10% | 9.30% | 9.15% | 8.17% | 7.02% |
|
|
|
|
|
|
8- He. bellottii Abacaxis | 23.18% | 19.99% | 9.86% | 9.51% | 9.47% | 8.37% | 3.98% |
|
|
|
|
|
9- He. bellottii Mamuru | 23.18% | 20.42% | 10.43% | 9.69% | 9.66% | 7.79% | 4.17% | 2.97% |
|
|
|
|
10- Hy. montagi | 22.66% | 18.08% | 18.88% | 18.43% | 17.87% | 16.40% | 16.63% | 18.26% | 17.85% |
|
|
|
11- Hy. mamuruensis | 21.12% | 17.31% | 15.83% | 14.79% | 15.26% | 14.28% | 14.97% | 16.80% | 15.98% | 9.33% |
|
|
12- Hy. herbertaxelrodi | 22.54% | 19.20% | 16.82% | 16.61% | 15.68% | 14.66% | 15.68% | 15.05% | 15.25% | 13.65% | 11.67% |
|
13- Hy. ericae | 22.29% | 17.70% | 15.92% | 16.97% | 16.02% | 14.79% | 16.41% | 16.19% | 17.22% | 12.35% | 11.16% | 9.56% |
Discussion
Hyphessobryconinae, one of the recently proposed subfamilies of Acestrorhamphidae, was proposed using only the species relationships recovered in Melo et al. (2024) phylogenomic study of Characidae. This subfamily lacks a morphological diagnosis, and it is divided into four main unnamed clades. Hyphessobrycon mamuruensis possesses two of the three typical features of species recovered in one of these four clades, specifically the one comprising most sampled species of the Hy. agulha and all sampled species of Hy. heterorhabdus species groups (the least inclusive clade with Hy. agulha and Hy. heterorhabdus, which we propose to be informally named as “Hy. agulha lineage”). These features are: an midlateral iridescent stripe and teeth with up to five cusps (Melo et al., 2024). Additionally, Hy. mamuruensis possesses a longitudinal narrow red stripe dorsally to the midlateral iridescent stripe as is typical for the species of the Hy. heterorhabdus species group (sensu Faria et al., 2021).
Our resulting tree, using sequences of the cytochrome c oxidase subunit I, recovers Hyphessobrycon mamuruensis as the sister species of Hy. montagi, with a genetic distance of 9.33% between them. Melo et al. (2024) did not sample neither of these species, however, in the present study, these species were recovered as a sister group of a clade containing Hy. ericae and Hy. herbertaxelrodi, studied by Melo et al. (2024), with moderate to strong bootstrap support (78%), suggesting that Hy. mamuruensis and Hy. montagi could be considered as part of the Hy. agulha lineage in the subfamily Hyphessobryconinae. Our molecular data also recover He. bellottii, Hy. heterorhabdus, Hy. eschwartzae, Hy. amapaensis,and Hy. sateremawe forming a well supported clade with 94% bootstrap support. This result agrees with that found by Melo et al. (2024) which found that He. cf. bellottii, Hy. heterorhabdus,and Hy. amapaensis belong to a lineage related with Hy. ericae and Hy. herbertaxelrodi.
The only congeners sharing a color pattern similar to Hyphessobrycon mamuruensis are Hy. hildae and Hy. taguae. Hyphessobrycon hildae is only known from its original description (Fernández-Yépez, 1950), and its holotype and only known specimen originated from the Río Autana, a tributary of the upper Río Orinoco basin in Venezuela, whereas Hy. taguae was described from localities in the Amazon and Orinoco basins in Colombia (García-Alzate et al., 2010). Additionally, a non-congener very similar to Hy. mamuruensis is Hemigrammus bellottii, a widely distributed species in the Amazon basin, and in fact both species were collected syntopically at the rio Mamuru. Both species share the presence of a conspicuous humeral blotch and the absence of any other conspicuous dark markings in the body or fins (compare Figs. 1 and 7). The diagnosis between Hy. mamuruensis from He. bellottii is essentially the same as its diagnosis from Hy. hildae and Hy. taguae, i.e.,the presence of a distinctly vertically elongated humeral blotch in Hy. mamuruensis, versus a humeral blotch that is only slightly higher than wide in He. bellottii, Hy. hildae, and Hy. taguae (Fig. 3). In fact, even though currently not congeneric with He. bellottii, no diagnosis among these three nominal species is available. Fernandez-Yépez (1950) noticed that Hy. hildae presents a tiny blotch at the base of the caudal fin, a feature we have observed in specimens identified by us as He. cf. bellottii from the upper Río Orinoco in Colombia. The systematics of these three nominal species is currently being addressed (FCTL, TCF, and C. A. García-Alzate, work in progress) and for the time being we propose to consider them as belonging to a putative Hemigrammus bellottii species-complex.
FIGURE 7| Hemigrammus bellottii color variation. Top: LBP 34285, 28.7 mm SL, Brazil, Pará, rio Trombetas basin near Floresta Estadual do Trombetas. Middle: LBP 34342, 37.2 mm SL, Brazil, Pará, igarapé tributary of rio Amazonas at Santarém-Mirim. Bottom: LBP 33135, 28.3 mm SL, Brazil, Pará, rio Mamuru basin.
Hyphessobrycon mamuruensis specimens used in this study were collected in the first ever ichthyological expedition to explore the rio Mamuru basin. There are six other main rivers in the same region, as rio Canumã, rio Abacaxis, rio Paraconi, rio Apoquitauá, rio Maués-Açu and rio Andirá, all of them presenting black water and discharging into the Paraná de Urariá and Paraná de Ramos. These represent two murky water channels, with the first one connecting the lower rio Madeira into the rio Amazonas and the second one connecting the eastern portion of the Paraná de Urariá to the rio Amazonas. The only other species previously described from this same region is Hy. sateremawe known from the rio Abacaxis and rio Maués-Açu basins. Some ichthyological surveys have been conducted at the rio Canumã, rio Abacaxis, rio Paraconi, rio Maués-Açu and rio Mamuru basins, but the whole area can still be considered sparsely sampled and consequently poorly known ichthyologically. Additional surveys in the area are necessary to fill this gap in the knowledge of Amazon fishes.
Comparative material examined. Hemigrammus bellottii: all from Brazil, river basins: Abacaxis: LBP 33007, 41, 16.6–28.4 mm SL. Amazonas (direct affluents):LBP 34342, 154, 20.7–36.8 mm SL; LBP 34476, 207, 18.9–33.1 mm SL; NMW 57254, 4, 24.6–29.8 mm SL, syntypes. Javari: ZUEC 16987, 206, 19.8–31.7 mm SL. Juruá: ZUEC 13717, 122, 16.5–31.5 mm SL. Madeira: ZUEC 7237, 39, 20.0–30.7 mm SL. Mamuru: LBP 33022, 1, 21.0 mm SL; LBP 33135, 1, 28.5 mm SL; LBP 33193, 15, 16.5–30.7 mm SL; LBP 33412, 46, 16.2–25.5 mm SL. Negro: ZUEC 18232, 45, 21.1–27.1 mm SL. Paraconi: MPEG 16045, 23, 27.4–33.1 mm SL. Solimões: ZUEC 15338, 145, 22.2–30.1 mm SL. Tefé: ZUEC 15360, 136, 20.1–32.3 mm SL. Trombetas: LBP 34285, 40, 23.8–30.4 mm SL. Urubu: ZUEC 11407, 5, 27.8–34.3 mm SL. Hemigrammus cf. bellottii: all from Colombia, river basins: Inirida: MPUJ 995, 18 of 35, 16.2–29.0 mm SL; MPUJ 1493, 7 of 13, 25.4–28.4 mm SL; MPUJ 1494, 4 of 7, 23.0–25.8 mm SL. Vichada: IAvH-P 10196, 7 of 15, 23.1–26.6 mm SL. Hyphessobrycon heterorhabdus: Brazil, rio Acará: LBP 35036, 411, 17.0–30.9 mm SL. Hyphessobrycon taguae: Colombia, rio Inirida: MPUJ 919, 10 of 49 paratypes, 24.3–26.3 mm SL.
Acknowledgments
André L. C. Canto, Frank R. V. Ribeiro and Carlison Silva provided logistical support. Fig. 1 was prepared with the help of Guilherme M. Dutra and Gabriel C. Deprá. FCTL is grateful to Carlos DoNascimiento (formerly at IAvH), Saul Prada-Pedreros and Alex Urbano-Bonilla (MPUJ), and Helmut Wellendorf and Ernst Mikschi (NMW) for allowing the examination of material under their care.
References
Edgar RC. Muscle: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004; 5:113. https://doi.org/10.1186/1471-2105-5-113
Faria TC, Lima FCT, Wosiacki WB. A new Hyphessobrycon (Characiformes: Characidae) from the Guiana Shield in northern Brazil. Copeia. 2020; 108(2):369–75. https://doi.org/10.1643/CI-19-311
Faria TC, Guimarães KLA, Rodrigues LRR, Oliveira C, Lima FCT. A new Hyphessobrycon (Characiformes: Characidae) of the Hyphessobrycon heterorhabdus species-group from the lower Amazon basin, Brazil. Neotrop Ichthyol. 2021; 19(1):e200102. https://doi.org/10.1590/1982-0224-2020-0102
Fernández-Yépez A. Algunos peces del río Autana. Novedades Cientificas, Ser Zool. 1950; 2:1–18.
Fink WL, Weitzman SH. The so-called cheirodontin fishes of Central America with descriptions of two new species(Pisces: Characidae). Smithson Contr Zool. 1974; 172:1–46. https://doi.org/10.5479/si.00810282.172
García-Alzate CA, Román-Valencia C, Taphorn DC. Two new species of Hyphessobrycon (Pisces: Characiformes: Characidae) from Putumayo River, with keys to the Colombian Hyphessobrycon heterorhabdus-group species. Brenesia. 2010; 70:33–40.
Géry J. Characoids of the World. Neptune City: TFH Publications; 1977.
Ingenito LF, Lima FCT, Buckup PA. A new species of Hyphessobrycon Durbin (Characiformes: Characidae) from the rio Juruena basin, Central Brazil, with notes on H. loweae Costa & Géry. Neotrop Ichthyol. 2013; 11(1):33–44. http://dx.doi.org/10.1590/S1679-6225201300010000
International Union for Conservation of Nature (IUCN). Standards and petitions committee. Guidelines for using the IUCN Red List categories and criteria [Internet]. Gland; 2024. Available from: http://www.iucnredlist.org/documents/RedListGuidelines.pdf
Ivanova NV, Dewaard JR, Hebert PDN. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes. 2006; 6(4):998–1002. https://doi.org/10.1111/j.1471-8286.2006.01428.x
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012; 28(12):1647–49. https://doi.org/10.1093/bioinformatics/bts199
Lima FCT, Coutinho DP, Wosiacki WB. A new Hyphessobrycon (Ostariophysi: Characiformes: Characidae) from the middle Amazon basin, Brazil. Zootaxa. 2014;3872(2):167–79. http://dx.doi.org/10.11646/zootaxa.3872.2.3
Lima FCT, Rodríguez-Olarte D, García-Alzate CA. A tale of false origins: Hyphessobrycon scholzei Ahl, 1937 and its junior synonym, Hyphessobrycon fernandezi Fernández-Yépez, 1972 (Teleostei: Characiformes: Acestrorhamphidae). Zootaxa. 2025a; 5594(1):50–60. https://doi.org/10.11646/zootaxa.5594.1.2
Lima FCT, Silva-Oliveira C, Oliveira C, Faria TC. A new Hyphessobrycon (Characiformes: Acestrorhamphidae) from the Central Amazon basin, Brazil. Pap Avulsos Zool. 2025b; 65:e202565011. https://doi.org/10.11606/1807-0205/2025.65.011
Melo BF, Ota RP, Benine RC, Carvalho FR, Lima FCT, Mattox GMT et al. Phylogenomics of Characidae, a hyper-diverse Neotropical freshwater fish lineage, with a phylogenetic classification including four families (Teleostei: Characiformes). Zool J Linn Soc. 2024; 202(1):zlae101. https://doi.org/10.1093/zoolinnean/zlae101
Mirande JM. Morphology, molecules and the phylogeny of Characidae (Teleostei, Characiformes).Cladistics. 2019; 35(3):282–300. https://doi.org/10.1111/cla.12345
Moreira CR, Lima FCT. Two new Hyphessobrycon (Characiformes: Characidae) species from Central Amazon basin, Brazil. Zootaxa. 2017;4318(1):123–34. http://dx.doi.org/10.11646/zootaxa.4318.1.5
Ohara WM, Lima FCT. Hyphessobrycon lucenorum (Characiformes: Characidae), a new species from the rio Madeira basin, Rondônia State, Brazil. Zootaxa. 2015; 3972(4):562–72. https://doi.org/10.11646/zootaxa.3972.4.7
Ohara WM, Teixeira TF, Albornoz-Garzón JG, Mirande JM, Lima FCT. Hyphessobrycon rheophilus, a new species from rapids of the Amazon and Orinoco river basins (Characiformes: Characidae: Stethaprioninae). Zootaxa. 2019;4712(4):561–75. http://dx.doi.org/10.11646/zootaxa.4712.4.5
Ota RR, Carvalho FR, Pavanelli CS. Taxonomic review of the Hyphessobrycon panamensis species-group (Characiformes: Characidae). Zootaxa. 2020; 4751(3):401–36.
http://dx.doi.org/10.11646/zootaxa.4751.3.1
Puillandre N, Brouillet S, Achaz G. ASAP: assemble species by automatic partitioning. Mol Ecol Resour. 2021; 21(2):609–20. https://doi.org/10.1111/1755-0998.13281
Sabaj MH. Codes for natural history collections in ichthyology and herpetology. Copeia.2020; 108(3):593–669. https://doi.org/10.1643/ASIHCODONS2020
Sabaj MH. Codes for natural history collections in ichthyology and herpetology (online supplement). 2023. Available from: https://www.asih.org/resources/standard-symbolic-codes
Taylor WR, Van Dyke GC. Revised procedures for staining and clearing small fishes and other vertebrates for bone and cartilage study. Cybium. 1985;9(2):107–19. Available from: https://sfi-cybium.fr/sites/default/files/pdfs-cybium/01-Taylor%5B92%5D107-119.pdf
Teixeira TF, Salvador GN, Mirande JM, Lima FCT. A new species of Psalidodon (Characiformes: Acestrorhamphidae) from the upper Rio São Francisco basin, Brazil, with comments on Hasemania. Ichthyol Herpetol. 2025; 113(1):131–42. https://doi.org/10.1643/i2024065
Toledo-Piza M, Baena EG, Dagosta FCP, Menezes NA, Andrade MC, Benine RC et al. Checklist of the species of the Order Characiformes (Teleostei: Ostariophysi). Neotrop Ichthyol. 2024; 22(1):e230086. https://doi.org/10.1590/1982-0224-2023-0086
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia’s fish species. Philos Trans R Soc B Biol Sci. 2005; 360(1462):1847–57. https://doi.org/10.1098/rstb.2005.1716
Weitzman SH, Palmer L. A new species of Hyphessobrycon (Teleostei: Characidae) from the Neblina region of Venezuela and Brazil, with comments on the putative ‘rosy tetra clade. Ichthyol Explor Freshw. 1997; 7(3):209–42.
Zhang J, Kapli P, Pavlidis P, Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013; 29(22):2869–76. https://doi.org/10.1093/bioinformatics/btt499
Authors
![]() Tiago C. Faria1
Tiago C. Faria1 ![]() ,
, ![]() Claudio Oliveira1,
Claudio Oliveira1, ![]() Iann Leonardo Pinheiro Monteiro2 and
Iann Leonardo Pinheiro Monteiro2 and ![]() Flávio César Thadeo de Lima3
Flávio César Thadeo de Lima3
[1] Laboratório de Biologia e Genética de Peixes (LBP), Instituto de Biociências de Botucatu (IBB), Universidade Estadual Paulista “Júlio de Mesquita Filho” (UNESP), 18618-970 Botucatu, SP, Brazil. (TCF) tc.faria@unesp.br (corresponding author), (CO) claudio.oliveira@unesp.br.
[2] Laboratório de Ictiologia, Museu Paraense Emílio Goeldi, Av. Perimetral, 1901, 66077-830 Belém, PA, Brazil. (ILPM) iannlpmonteiro@gmail.com.
[3] Museu de Diversidade Biológica, Universidade Estadual de Campinas, Caixa Postal 6109, 13083-863 Campinas, SP, Brazil. (FCTL) fctlima@gmail.com.
Authors’ Contribution 

Tiago C. Faria: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing-original draft.
Claudio Oliveira: Data curation, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing-review and editing.
Iann Leonardo Pinheiro Monteiro: Methodology.
Flávio César Thadeo de Lima: Conceptualization, Data curation, Investigation, Supervision, Writing-review and editing.
Ethical Statement
No experiments involving live animals were conducted for this study. All specimens were obtained through field collection under the permanent license for biological material collection issued to Claudio Oliveira (SISBIO license number 13843-5).
Competing Interests
The author declares no competing interests.
Data availability statement
The authors confirm that the molecular data supporting the findings of this study are available in Genbank and/or BOLD with access numbers available in Tab. 1. Morphological data and distributions are available within the article.
Funding
The authors were funded by Fundação de Amparo à Pesquisa do Estado de São Paulo – FAPESP (TCF, grant #2021/00242–0; CO, grant #2020/13433–6; FCTL, grants #2011/51532-7 and 2013/20936–0), Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq (CO, grants #306054/2006–0 and #441128/2020–3) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001. All specimens of Hyphessobrycon mamuruensis were obtained during an expedition funded partly by the “Projeto Filogenia e identificação molecular de peixes da superordem Ostariophysi (Chordata: Actinopterygii) utilizando abordagens genômicas” (FAPESP grant #2020/13433–6) and partly by the first author project “Sistemática e filogeografia dos tetras do complexo Hyphessobrycon pulchripinnis (Characiformes: Characidae)” (FAPESP grant #2021/00242–0).
How to cite this article
Faria TC, Oliveira C, Monteiro ILP, Lima FCT. A new Hyphessobrycon (Characiformes: Acestrorhamphidae) from the rio Mamuru basin, Amazonas State, Brazil. Neotrop Ichthyol. 2025; 23(3):e240073. https://doi.org/10.1590/1982-0224-2024-0073
Copyright
This is an open access article under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in any medium, provided the original work is properly cited.
Distributed under
Creative Commons CC-BY 4.0

© 2025 The Authors.
Diversity and Distributions Published by SBI
![]() Accepted June 10, 2025
Accepted June 10, 2025
![]() Submitted July 29, 2024
Submitted July 29, 2024
![]() Epub September 29, 2025
Epub September 29, 2025