![]() Rafael B. de Almeida1,
Rafael B. de Almeida1, ![]() Matheus Azambuja1,
Matheus Azambuja1, ![]() Viviane Nogaroto2,
Viviane Nogaroto2, ![]() Claudio Oliveira3,
Claudio Oliveira3, ![]() Fábio F. Roxo3,
Fábio F. Roxo3, ![]() Cláudio H. Zawadzki4 and
Cláudio H. Zawadzki4 and ![]() Marcelo R. Vicari1,2
Marcelo R. Vicari1,2 ![]()
PDF: EN XML: EN | Cite this article
Abstract
Isbrueckerichthys is a genus of armored catfish (Siluriformes: Loricariidae) with five species in the lowlands from the Ribeira de Iguape basin and in the uplands of the Tibagi River basin. Despite the validation of the morphological species, molecular data to investigate gene flow and species delimitation have not been completed for all species. For this purpose, we compared sequences of COI region associated with barcoding molecular identification, aiming for a species delimitation analysis and generating population data of I. alipionis, I. epakmos, I. duseni, I. cf. duseni, I. saxicola, and I. calvus. The K2P genetic distance, molecular species delimitation analysis, and well-sustained branches in the phylogenetic tree validate I. alipionis, I. epakmos, and I. duseni, and suggest I. cf. duseni as a valid molecular operational taxonomic unit. However, no differences were detected between I. saxicola and I. calvus. The discordance between molecular and morphological species may be due to the recent dispersal of some Isbrueckerichthys representatives at the border between the Ribeira de Iguape and Tibagi basins. The isolation features of the mountainous region of Ribeira de Iguape basin and headwaters captures to uplands are presented to explain the dispersion and the cases of incipient speciation in Isbrueckerichthys lineages.
Keywords: Incipient speciation, Integrative taxonomy, Molecular operational taxonomic units, Molecular species delimitation, Systematics.
Isbrueckerichthys é um gênero de cascudinhos (Siluriformes: Loricariidae) que reúne cinco espécies morfológicas nas terras baixas da bacia do Ribeira de Iguape e nas terras altas da bacia do rio Tibagi. Apesar da validação das espécies morfológicas, estudos moleculares não tinham sido realizados para investigar o fluxo gênico e delimitação molecular das espécies. Para tanto, comparamos sequências da região COI associadas à identificação molecular por código de barras visando uma análise de delimitação de espécies e dados populacionais entre I. alipionis, I. epakmos, I. duseni, I. cf. duseni, I. saxicola e I. calvus. Os dados de distância genética K2P, a análise de delimitação molecular de espécies e ramos bem sustentados na árvore filogenética validam I. alipionis, I. epakmos e I. duseni, e sugere I. cf. duseni como unidade taxonômica operacional molecular. Ainda, não foram observadas diferenças entre I. saxicola e I. calvus. A discordância entre espécies moleculares e morfológicas pode ser devida à recente dispersão e diversificação de alguns representantes de Isbrueckerichthys na fronteira das bacias de Ribeira de Iguape e Tibagi. Características de isolamento da região serrana da bacia do Ribeira de Iguape e eventos de capturas de cabeceiras para terras altas foram utilizadas para explicar a dispersão e os casos de especiação incipiente em algumas linhagens de Isbrueckerichthys.
Palavras-chave: Delimitação molecular de espécies, Especiação incipiente, Sistemática, Taxonomia integrativa, Unidade taxonômica operacional molecular.
Introduction
Loricariidae is one of the most species-rich fish families, with 1,056 valid species grouped in 115 genera (Fricke et al., 2024). The broad species diversification in Loricariidae makes it difficult for accurate determination of species relationships, leading to doubts in subfamilies, tribes, and genera (Isbrücker, 1980; Armbruster, 2004; Reis et al., 2006; Lujan et al., 2015; Covain et al., 2016; Pereira, Reis, 2017; Roxo et al., 2019). Another crucial problem in Loricariidae is the taxonomic uncertainties that occur in paired species belonging to isolated hydrographic basins (Anjos et al., 2021; de Sousa et al., 2021; Lustosa-Costa et al., 2022) or species resident in tributaries and with low vagility (Albert et al., 2020; de Sousa et al., 2021).
The genus Isbrueckerichthys (Loricariidae: Siluriformes) was proposed by Derijst (1996), justified on the discovery of an earlier type-species designation by Regan (1920), where Pareiorhaphis duseni (Miranda Ribeiro, 1907) and Pareiorhaphis alipionis (Gosline, 1947) were moved to the new genus. Their representatives are small-sized species of armored catfishes distinct from the sister taxon Neoplecostomus Eigenmann & Eigenmann, 1888 by several diagnostic morphological characteristics, such as a small naked area behind the pterotic-supracleithrum, abdomen with small platelets embedded in the skin between pectoral girdle and pelvic-fin insertions, one spine and seven branched rays in the dorsal fin, caudal peduncle ovoid in cross-section, and lacking a series of papillae on the lower lip of the dentaries (Derijst, 1996; Pereira, Oyakawa, 2003; Jerep et al., 2006). Isbrueckerichthys species inhabit headwater streams, usually rapids (<1 m deep), with well-oxygenated water and substrates consisting of rocks (Pereira, Reis, 2002). The first species recognized as Isbrueckerichthys (I. duseni, I. alipionis, and I. epakmos Pereira & Oyakawa, 2003)occur in the plain coastal lands of the Atlantic and are restricted to the Ribeira de Iguape River basin (Derijst, 1996; Pereira, Oyakawa, 2003; Dagosta et al., 2024). Afterward, Jerep et al. (2006) described two species (I. saxicola Jerep, Shibatta, Pereira & Oyakawa, 2006and I. calvus Jerep, Shibatta, Pereira & Oyakawa, 2006) from upland rivers of the Tibagi basin, belonging to the upper Paraná River.
For some authors, a taxonomic system based strictly on morphology underestimates the cryptic species diversity, which is common in many groups (Knowlton, 1993; Jarman, Elliott, 2000; Hebert et al., 2003). The essence of this limitation is the phenotypic plasticity and the genetic variability in the characters that can lead to erroneous identifications (Hebert et al., 2003). In Neotropical fishes, molecular analyses involving species delimitation or population biology have contributed to the understanding of distribution, taxonomy, and systematics in several groups (do Nascimento et al., 2018; Santos et al., 2019; Argolo et al., 2020; Herrera-Collazos et al., 2020; Traldi et al., 2020; Ariza et al., 2022; Azambuja et al., 2022; Silva-Santos et al., 2023; Souza et al., 2023). Previous studies have stipulated parameters for standardizing values for genetic divergence for molecular markers utilized for species identification (Ward, 2009). For several authors, the interspecific divergence must be ten times higher than the intraspecific divergence for a taxa validation (Hebert et al., 2004). Ward (2009) proposed that fish specimens that are strictly related and present genetic divergences above 2% belong to different species. Although the genetic divergence values associated with molecular markers used for molecular identification are standardized for some organisms, it is known that the trigger values for intra/interspecific divergence are variable and are influenced by the variation in the substitution rate through the lineages (Barraclough et al., 2009).
In this context, integrative taxonomy/phylogenetics is a contemporary strategy addressed to questions in which single data-based approaches reveal less consistent conclusions rather than those by combined multisource evidence in terms of detecting the species richness within supposedly homogeneous taxa (Padial et al., 2010; Schlick-Steiner et al., 2010; Grković et al., 2017). Distinct species delimitation methods, such as ABGD, ASAP, and bPTP, have been proposed to analyze a single locus (Pons et al., 2006; Zhang et al., 2013). In general, these analyses have demonstrated that the combination of different methods, as well as integrative taxonomy, allows more representative results for the identification of lineages in species (Tang et al., 2014; Kapli et al., 2017; Luo et al., 2018; Serrano et al., 2018; Cardoso et al., 2023; Souza et al., 2023). Isbrueckerichthys is a Loricariidae genus with reformulations and species descriptions in the last three decades (Derijst, 1996; Pereira, Oyakawa, 2003; Jerep et al., 2006). However, molecular species delimitation studies are lacking in the genus. Therefore, this study aimed to conduct a molecular delimitation analysis in all Isbrueckerichthys species inhabiting the lowlands and uplands in south and southeast Brazilian drainages.
Material and methods
Biological samples. Specimens of Isbrueckerichthys were collected in different tributaries of the lowlands from the Ribeira de Iguape basin and in tributaries of the uplands from the Tibagi basin (Fig. 1). Specimens were deposited as vouchers in the ichthyological collection at Núcleo de Pesquisas em Limnologia, Ictiologia e Aquicultura of Universidade Estadual de Maringá, Maringá (NUP) and in the Laboratório de Biologia e Genética de Peixes of the Universidade Estadual Paulista “Júlio de Mesquita Filho”, Botucatu (LBP) (Tab. 1).
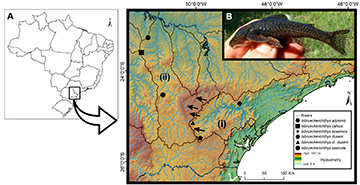
FIGURE 1| Collection sites and representative image of an Isbrueckerichthys species. A. Partial map of Brazil highlighting south/southern hydrographic basins of the Ribeira de Iguape River (i) and Tibagi River (ii). The arrows show the Ponta Grossa Arch region in eastern Paraná State, Brazil (a relief barrier between lowlands and uplands hydrographic basins). The map shows elevation and valleys in the relief, and the geometric shapes indicate the collection site of the specimens of Isbrueckerichthys analyzed. B. Live specimen of I. calvus photograph taken during capture in the Juruba stream.
TABLE 1 | Collection sites of the Isbrueckerichthys species, number of individuals analyzed, and voucher numbers.
Species | Number of individuals | Samples | Coordinates | Voucher number |
Isbrueckerichthys alipionis | 7 | Betari River/Ribeira de Iguape basin | 24°33’42.1”S 48°40’05.7”W | LBP 7373 |
Isbrueckerichthys calvus | 5 | Juruba stream (type- locality)/Tibagi basin | 23°34’24.68”S 51°23’50.30”W | LBP 6404 |
Isbrueckerichthys duseni | 5 | Açungui River/Ribeira de Iguape basin |
25°22’43.9”S
| NUP 16200 |
Isbrueckerichthys cf. duseni | 8 | D’Areia stream/Tibagi basin |
25°14’09.0”S
| NUP 20293 |
Isbrueckerichthys epakmos | 3 | Juquiá River/ Ribeira de Iguape basin |
24°01’25.2”S
| LBP 7385 |
Isbrueckerichthys saxicola | 5 | Jacutinga stream (type- locality)/Tibagi basin | 23°14’29.87”S 51°13’5.10”W | LBP 40259-40263 |
3 | Charqueada River/Tibagi basin |
24°29’09.0”S
| NUP 20291 |
Molecular analyses. Genomic DNA of thirty-one individuals of Isbrueckerichthys were extracted from liver samples using the CTAB (cetyltrimethylammonium bromide) method (Murray, Thompson, 1980). The barcode region of the mitochondrial gene cytochrome c oxidase subunit I (COI) was amplified by PCR using the primers Fish F1 and Fish R1 (Ward et al., 2005). The reaction was composed of 1x Taq Reaction buffer (200 mM Tris pH 8.4, 500 mM KCl), 0.2 mM dNTPs, 1 mM MgCl2, 1 U of Taq DNA polymerase (Invitrogen), 0.4 µM of each primer, and 40 ng of DNA. The following reaction program was used: initial denaturation for 10 min at 94 °C, 35 cycles of 94 °C for 1 min, 51.8 °C for 45 sec and 72 °C for 1 min, and a final extension at 72 °C for 10 min. PCR products were purified with the Illustra GFX PCR DNA and Gel Band Purification (GE Healthcare) and sequenced in an ABI-Prism 3500 Genetic Analyzer (Applied Biosystems).
Sequences were analyzed using Geneious v. 7.1.9 (Kearse et al., 2012) software and submitted to GenBank. Five sequences of I. saxicola (GU701901 – GU701903; GU701905 – GU701906) from the Jacutinga River were recovered from the GenBank. The sequences were aligned with the ClustalW algorithm integrated into the Geneious program. Nucleotide (π) and haplotype (Hd) diversity were computed by DnaSP v. 5 (Librado, Rozas, 2009). Sequences were separated into groups, and genetic distances were calculated in MEGA X (Kumar et al., 2018) under the Kimura-2-Parameters evolution model and 1,000 bootstrap replications. A haplotype network was generated through the median-joining criterion (Bandelt et al., 1999) in the PopArt v. 1.7 software (Leigh, Bryant, 2015). Structural analysis was performed by assigning each individual to the respective populations using Bayesian Analyses of Population Structure – BAPS 6, allowing for a range of 1–6 (Corander et al., 2004, 2008). Population structuring and analysis of molecular variance (AMOVA) (Excoffier et al., 1992) were performed by Arlequin v. 3.5.2.2 software (Excoffier, Lischer, 2010).
One Pareiorhaphis splendens (Bizerril, 1995) (Neoplecostomini) COI sequenceEU359449.1 was aligned to Isbrueckerichthys with ClustalW and used to root trees, according to previous molecular phylogenetic studies (Roxo et al., 2012). The alignment was submitted to jModelTest 2 (Darriba et al., 2012) using the corrected Akaike information criterion (AICc) to select the best-fit nucleotide evolution HKY model for downstream analyses. A Bayesian inference tree was generated in the MrBayes 3.2 program (Huelsenbeck, Ronquist, 2001; Ronquist, Huelsenbeck, 2003), applying 100,000,000 interactions of MCMC, sampling trees every 10,000 generations and burn-in of 10,000,000.
Three methods for species delimitation were applied: Automatic Barcode Gap Discovery (ABGD) (Puillandre et al., 2012), Assemble Species by Automatic Partitioning (ASAP) (Puillandre et al., 2020), and Bayesian Poisson Tree Processes (bPTP) (Zhang et al., 2013). The alignment generated for the sequences was used as an input file for the delimitation of species in the ABGD web server (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html) and in the ASASP web server (https://bioinfo.mnhn.fr/abi/public/asap/asapweb.html), the Kimura (K80) TS/TV was selected for distance mode and the other parameters were kept at default. For bPTP analysis, the Bayesian tree generated in MrBayes was used as an input file in a bPTP web server (https://species.h-its.org/ptp/). There were 500,000 MCMC generations run (thinning = 500), and the other parameters were kept at default.
Results
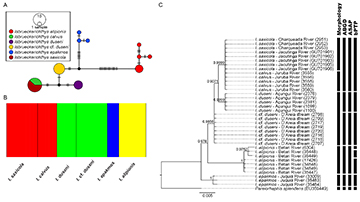
Sequences of the COI gene were obtained from the Isbrueckerichthys specimens and deposited in GenBank (PP391830–PP391860). All sequences were of high quality and presented no evidence of insertions, deletions, or stop codons. The obtained sequences were aligned with the sequences of I. saxicola from the GenBank database, and a matrix of 36 sequences with 612-bp was generated. The nucleotide diversity (π) was 0.01369, and haplotype diversity (Hd) was 0.806. A total of ten haplotypes were observed: Isbrueckerichthys alipionis had four haplotypes; I. calvus and I. saxicola shared a unique haplotype; individuals of I. duseni and I. cf. duseni had one exclusive haplotype to each species; and in I. epakmos three haplotypes were observed(Fig. 2A).
FIGURE 2| Molecular data of Isbrueckerichthys specimens. A. Haplotype network showing the relationship among the COI sequences. B. Structural inference generated by BAPs (K = 4). C. Bayesian inference tree showing the phylogenetic relationship of Isbrueckerichthys sequences (numbers on the branches correspond to the posterior probability). Vertical black bars correspond to the morphological classification of the specimens, and the molecular units determined using ABGD, ASAP, and bPTP species delimitation methods.
The intraspecific distance for each group ranged from 0 to 0.23%, and the interspecific distance ranged from 0 to 3.63% (Tab. 2). The BAPs results indicated an optimal partition of four clusters (k = 4; marginal log-likelihood = -240.4536). Individuals of I. calvus and I. saxicola were assigned as a single group, I. duseni,and I. cf. duseni were separated into a second cluster, and I. epakmos and I. alipionis represent independent groups (Fig. 2B). The AMOVA analysis, considering the clusters delimited by BAPs, shows a variance value of 83.98% among groups (Tab. 3), with an FST value of 0.96692 (p < 0.05). The pairwise FST values between each population ranged from 0.00 to 1.00 (Tab. 4).
TABLE 2 | Estimates of evolutionary divergence over sequence pairs among groups and standard error in the Isbrueckerichthys species. Bold values in main diagonal show the intraspecific genetic distance. The number of base substitutions per site from averaging over all sequence pairs between groups is shown. Analyses were conducted using the Kimura 2-parameter model. This analysis involved 36 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 612 positions in the final dataset.
| I. alipionis | I. calvus | I. duseni | I. cf. duseni | I. epakmos | I. saxicola |
I. alipionis | 0.0023 (0.0012) |
|
|
|
|
|
I. calvus | 0.0301 (0.0072) | 0.0000 (0.0000) |
|
|
|
|
I. duseni | 0.0336 (0.0077) | 0.0066 (0.0033) | 0.0000 (0.0000) |
|
|
|
I. cf. duseni | 0.0283 (0.0070) | 0.0049 (0.0029) | 0.0049 (0.0027) | 0.0000 (0.0000) |
|
|
I. epakmos | 0.0363 (0.0078) | 0.0194 (0.0055) | 0.0194 (0.0054) | 0.0144 (0.0045) | 0.0022 (0.0016) |
|
I. saxicola | 0.0301 (0.0072) | 0.0000 (0.0000) | 0.0066 (0.0033) | 0.0049 (0.0029) | 0.0194 (0.0055) | 0.0000 (0.0000) |
TABLE 3 | Analysis of molecular variance (AMOVA) from the Isbrueckerichthys species. Four groups were considered: I. alipionis; I. calvus + I. saxicola; I. epakmos; I. duseni + Isbrueckerichthys cf. duseni. Bold values were significant (p < 0.05).
Source of variation |
d.
| Sum of squares | Variance components | Percentage of variation |
Among groups | 3 | 131.761 | 4.75474 | 83.98 |
Among populations within groups | 2 | 9.231 | 0.71956 | 12.71 |
Within populations | 30 | 5.619 | 0.18730 | 3.31 |
Total | 35 | 146.611 | 5.66160 | 100 |
Fixation Indices FSC: 0.79346 FST: 0.96692 FCT: 0.83982 |
|
|
|
|
TABLE 4 | Pairwise FST values among populations of Isbrueckerichthys (lower diagonal) and p-values (upper diagonal). Bold values were significant (p < 0.05).
| I. alipionis | I. calvus | I. duseni | I. cf. duseni | I. epakmos | I. saxicola |
I. alipionis | – | 0.0000+-0.0000 | 0.0000+-0.0000 | 0.0000+-0.0000 | 0.0000+-0.0000 | 0.0000+-0.0000 |
I. calvus | 0.9523 | – | 0.0180+-0.0121 | 0.0090+-0.0091 | 0.0180+-0.0121 | 0.9910+-0.0030 |
I. duseni | 0.9571 | 1.0000 | – | 0.0090+-0.0091 | 0.0090+-0.0091 | 0.0000+-0.0000 |
I. cf. duseni | 0.9608 | 1.0000 | 1.0000 | – | 0.0000+-0.0000 | 0.0000+-0.0000 |
I. epakmos | 0.9345 | 0.9615 | 0.9615 | 0.9649 | – | 0.0090+-0.0091 |
I. saxicola | 0.9630 | 0.0000 | 1.0000 | 1.0000 | 0.9741 | – |
The Bayesian phylogenetic tree revealed five supported clades: I. epakmos, I. alipionis, Isbrueckerichthys cf. duseni, I. duseni, and I. calvus + I. saxicola (Fig. 2C). Species delimitation methods returned different results (Fig. 2C). The ABGD method resulted in seven partitions ranging from two to nine species, with partition 4 (prior maximal distance P = 0.004642) indicating six species, including the outgroup. ABGD recognized I. epakmos, I. alipionis, I. duseni,and Isbrueckerichthys cf. duseni as single groups and grouped I. calvus and I. saxicola. The best partition identified by the ASAP method (ASAP-score: 3.50; p-value: 1.74e-02; W: 3.13e-05; Threshold dist.: 0.004094) recognized the same six species as the ABGD method. The ML solution of bPTP delimited eight species, including the outgroup. bPTP separated I. alipionis into three species and grouped I. calvus and I. saxicola.
Discussion
Evolutionary biologists agree that species are lineages or metapopulations that evolve separately, with disagreements remaining only about where, along the continuum of divergence, separate lineages should be considered distinct species (Wiley, 1978; Hey, 2006; Mallet, 2008). Dias Filho et al. (2020) argued that molecular techniques should complement classical taxonomic identification for a detailed description of organisms. Data analysis showed a restricted relationship among Isbrueckerichthys species, with some divergences concerning morphological identification for the samples obtained in the delimitations between the First (lowlands in Ribeira de Iguape basin) and the Second (uplands in Tibagi basin) Paraná State plateaus, Brazil.
The data obtained from COI sequences analysis demonstrated that I. epakmos, I. alipionis, and I. duseni, all inhabitants of the Ribeira de Iguape River basin, present K2P divergence values greater than or close to 2%, well-supported branches in the phylogenetic tree, and pairwise Fst close to 1, corroborating to the absence of gene flow and the occurrence of the nominal taxa. The Ribeira de Iguape River basin is in an intricate mountainous region with high isolating potential for aquatic organisms (Ribeiro, 2006), and Roxo et al. (2012) proposed that the ancestor of Isbrueckerichthys inhabited tributaries from this basin at least 20 million years ago. Langeani (1990) stated that most Neoplecostomini (Neoplecostominae in that work) species have low vagility and difficulties adapting to streams that occur at low altitudes due to the low oxygen concentration in the water compared to rivers in higher regions. Thus, streams in valleys between mountains (low altitude) could hinder the dispersal of Neoplecostomini species into different tributaries of the Ribeira de Iguape River basin (Fig. 1). These geological and ecological factors and evolutionary times allow for congruence between the molecular and morphological divergence data for I. epakmos, I. alipionis, and I. duseni.
The methods for species delimitation included I. cf. duseni (D’Areia stream), sampled in the border of the Ribeira de Iguape basin, the limit with Tibagi basin in Paraná State, as a new taxon for Ribeira de Iguape basin. Comparing specifically I. duseni vs. I. cf. duseni, 0.49% of K2P genetic distance was observed. However, the pairwise Fst showed gene flow restriction, and branches well-supported in the phylogenetic tree topology for I. duseni vs. I. cf. duseni were shown. Some Neotropical fish species demonstrate distinct taxonomic entities with genetic distances lower than 2%, although they are arranged in monophyletic groups (Bellafronte et al., 2013; Traldi et al., 2020). Based on molecular data, several studies have highlighted cryptic diversity for neotropical freshwater fish (Melo et al., 2016; Guimarães et al., 2019; Mateussi et al., 2019; Souza et al., 2021, 2023; Cardoso et al., 2023). In addition, as mentioned above, the specimens of Isbrueckerichthys could have gene flow restriction due to their biological features associated with the geological formation of the Ribeira de Iguape tributaries. There is a valley between the sample localities of I. duseni and I. cf. duseni (Fig. 1), which, according to Langeani (1990), makes dispersal difficult and, in this case, could contribute to genetic isolation. The data suggests that a molecular operational taxonomic unit for I. cf. duseni in the Ribeira de Iguape basin or a case of incipient speciation should be considered.
The genetic comparison among I. duseni, I. cf. duseni, I. saxicola,and I. calvus also demonstrated low K2P genetic distance (0.49–0.66%), high Fst, and branches well-supported in the phylogenetic tree. Despite low K2P values, I. duseni and Isbrueckerichthys cf. duseni are geographically isolated from I. saxicola and I. calvus by the Ponta Grossa Arch, an uplift of the relief that isolated the Ribeira de Iguape basin from upland rivers (Fig. 1), such as Tibagi basin (Ribeiro, 2006). Isbrueckerichthys duseni and I. cf. duseni were considered valid taxa in species delimitation methods, but a morphological and molecular data incongruence was observed between I. saxicola and I. calvus. These two species were described from the Tibagi basin by Jerep et al. (2006) having morphological differences with congeners including: bicuspid teeth, hypertrophied odontodes, and a more extended spine in the pectoral fin. The morphological differentiation between I. calvus and I. saxicola is based on the number of odontodes on the abdominal platelet (six or more in I. saxicola vs. up to six in I. calvus), by the absence/presence of a flattened area in the inferior portion of the two first plates of the lateral line, by the degree of exposition of the cleithrum and by the degree of convexity and exposition of the supraoccipital plane (Jerep et al., 2006). Although the morphological description of Jerep et al. (2006) is well-detailed, the morphological divergence is not accompanied by a genetic divergence. All methods based on the molecular species delimitation used in this study, using a single locus analysis, agree with I. saxicola and I. calvus as a single taxon. In addition, a pairwise Fst 0 was estimated between the localities of the I. saxicola and I. calvus samples, indicating no population structure.
In turn, it has been proposed that recent speciation in newly inhabited areas may present an intermediary stage of speciation due to the short space of evolutionary time for diversification, especially cases of incomplete lineage sorting and lack of divergence (Roxo et al., 2012, 2014, 2017). Neoplecostomini species occurring in the uplands of the Tibagi basin, like the case of Isbrueckerichthys, were proposed to inhabit this new area from dispersion of fishes through headwaters capture events between the coastal regions and those more elevated in the borders of the crystalline shield (Ribeiro, 2006; Roxo et al., 2012, 2014; Dagosta et al., 2024). Roxo et al. (2012) also suggested that the dispersal occurred around 4.8 (2.3–7.8), 5.9 (2.9–9.5), and 10.6 (5.5–16.5) million years ago. Based on this information, it is possible to verify that the morphological diversification between I. calvus and I. saxicola is not accompanied by a rate of nucleotide substitution, probably due to the short time of dispersal and distribution of Isbrueckerichthys in the Tibagi River basin.
Even with the emergence of combined multiple data sets strategies applied to diversity analysis, according to some authors, it is a challenge to evaluate species delimitation in considering demes that comprise geographically widespread series of similar individuals (de Queiroz, 2007; Willis, 2017), or those that emerged from a short period of dispersal in evolutionary diversification. However, also is expected a degree of phenotypic (morphological, ecological, and behavioral) divergence among sub-populations due to genetic drift and local adaptation (López-Fernández et al., 2005; de Queiroz, 2007; Zapata, Jiménez, 2011; Willis et al., 2017). At this point, the morphological differences between I. calvus and I. saxicola could be explained by their dispersal in distinct tributaries (environments) in the Tibagi basin. To solve these cases of incomplete lineage sorting and lack of divergence, as here demonstrated in Isbrueckerichthys,some authors argue that a multilocus analysis, phylogenomics methods, and multiple datasets are important for both phylogenetic resolution and in-depth investigation of speciation (López-Fernández et al., 2005, 2010; Ilves et al., 2018).
Integrative studies of molecular and morphological investigations may be considered for species delimitation because they represent complementary approaches practical for the group’s phylogeny, systematics, evolution, and phylogeography. However, groups with recent diversification (as revealed by similar or identical genetics coupled with morphological differentiation) still challenge integrative taxonomy. Here, there is low genetic divergence among Isbrueckerichthys that inhabit the border of Ribeira de Iguape basin in Paraná State and those from uplands in the Tibagi basin, which can be considered recent speciation cases. In I. calvus and I. saxicola in the Tibagi basin, molecular analysis indicates that morphological diversification between the species is not accompanied by genetic diversification.
Acknowledgments
MRV received financial support from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 313566/2023–2) and Fundação Araucária de Apoio ao Desenvolvimento Científico e Tecnológico do Estado do Paraná (009–2016). RBA and MA received financial support from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES – Finance Code 001). CO received financial support from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP grant 2020/13433–6) and CNPq (processes 306054/2006–0 and 441128/2020–3).
References
Albert JS, Tagliacollo VA, Dagosta F. Diversification of Neotropical freshwater fishes. Annu Rev Ecol Evol Syst. 2020; 51:27–53. https://doi.org/10.1146/annurev-ecolsys-011620-031032
Anjos MS, Queiroz LJ, Penido IS, Bitencourt JA, Barreto SB, Sarmento-Soares LM, Batalha-Filho H, Affonso PRAM. A taxonomically complex catfish group from an underrepresented geographic area: systematics and species limits in Hypostomus Lacépède, 1803 (Siluriformes, Loricariidae) from Eastern South America. J Zoolog Syst Evol Res. 2021; 59(8):1994–2009. https://doi.org/10.1111/jzs.12572
Argolo LA, López-Fernández H, Batalha-Filho H, Affonso PRAM. Unraveling the systematics and evolution of the ‘Geophagus’ brasiliensis (Cichliformes: Cichlidae) species complex. Mol Phylogenet Evol. 2020; 150:106855. http://dx.doi.org/10.1016/j.ympev.2020.106855
Ariza AA, Adachi AMCL, Roque P, Hazin FHV, Vianna M, Rotundo MM, Delpiani SM, Astarloa JMD, Delpiani G, Oliveira C, Foresti F, Cruz VP. DNA Barcoding and species delimitation for dogfish sharks belonging to the Squalus genus (Squaliformes: Squalidae). Diversity. 2022; 14(7):544. http://dx.doi.org/10.3390/d14070544
Armbruster JW. Phylogenetic relationships of the suckermouth armoured catfishes (Loricariidae) with emphasis on the Hypostominae and the Ancistrinae. Zool J Linn Soc. 2004; 141(1):1–80. http://dx.doi.org/10.1111/j.1096-3642.2004.00109.x
Azambuja M, Marcondes DS, Nogaroto V, Moreira-Filho O, Vicari MR. Population structuration and chromosomal features homogeneity in Parodon nasus (Characiformes: Parodontidae): a comparison between Lower and Upper Paraná River representatives. Neotrop Ichthyol. 2022; 20(1):e210162. http://dx.doi.org/10.1590/1982-0224-2021-0162
Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999; 16(1):37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036
Barraclough TG, Hughes M, Ashford-Hodges N, Fujisawa T. Inferring evolutionarily significant units of bacterial diversity from broad environmental surveys of single-locus data. Biol Lett. 2009; 5(3):425–28. https://doi.org/10.1098/rsbl.2009.0091
Bellafronte E, Mariguela TC, Pereira LHG, Oliveira C, Moreira-Filho O. DNA barcode of Parodontidae species from the La Plata river basin – applying new data to clarify Taxonomic problems. Neotrop Ichthyol. 2013; 11(3):497–506. https://doi.org/10.1590/S1679-62252013000300003
Cardoso VC, Dutra GM. Description of a new species of glass knifefish genus Eigenmannia (Gymnotiformes: Sternopygidae) from the upper rio Paraná basin, based on anatomical, karyotypic, and molecular evidences. Neotrop Ichthyol. 2023; 21(4):230090. https://doi.org/10.1590/1982-0224-2023-0090
Corander J, Marttinen P, Sirén J, Tang J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics. 2008; 9:539. https://doi.org/10.1186/1471-2105-9-539
Corander J, Waldmann P, Marttinen P, Sillanpää MJ. BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics. 2004; 20(15):2363–69. https://doi.org/10.1093/bioinformatics/bth250
Covain R, Fisch-Muller S, Oliveira C, Mol JH, Montoya-Burgos JI, Dray S. Molecular phylogeny of the highly diversified catfish subfamily Loricariinae (Siluriformes, Loricariidae) reveals incongruences with morphological classification. Mol Phylogenet Evol. 2016; 94:492–517. http://dx.doi.org/10.1016/j.ympev.2015.10.018
Dagosta FCP, Monção MS, Nagamatsu BA, Pavanelli CS, Carvalho FR, Lima FCT, Langeani F, Dutra GM, Ota RR, Seren TJ, Tagliacollo V, Menezes NA, Britski HA, de Pinna M. Fishes of the upper rio Paraná basin: diversity, biogeography and conservation. Neotrop Ichthyol. 2024; 22(1):e230066. https://doi.org/10.1590/1982-0224-2023-0066
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012; 9:772. https://doi.org/10.1038/nmeth.2109
Derijst E. Note on the type species of the mailed catfish genus Hemipsilichthys Miranda Ribeiro, 1918 (Pisces: Siluriformes; Loricariidae), with the introduction of Isbrueckerichthys nom. nov. Aquarium Wereld. 1996; 49:62–64.
Dias Filho CR, Rodrigues EL, Malaghini M, Francez PAC, Garrido RG. Introdução à genética forense. Campinas: Millennium Editora; 2020.
Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010; 10(3):564–67. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992; 131(2):479–91. https://doi.org/10.1093/genetics/131.2.479
Fricke R, Eschmeyer WN, van der Laan R. Eschmeyer’s catalog of fishes: genera, species, references [Internet]. San Francisco: California Academy of Science; 2024. Available from: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp
Grković A, Vujić A, Chroni A, van Steenis J, Ðan M, Radenković S. Taxonomy and systematics of three species of the genus Eumerus Meigen, 1822 (Diptera: Syrphidae) new to southeastern Europe. Zool Anz. 2017; 270:176–92. https://doi.org/10.1016/j.jcz.2017.10.007
Guimarães EC, Brito PS, Feitosa LM, Costa LFC, Ottoni FP. A new cryptic species of Hyphessobrycon Durbin, 1908 (Characiformes, Characidae) from the eastern Amazon, revealed by integrative taxonomy. Zoosyst Evol. 2019; 95(2):345–60. https://doi.org/10.3897/zse.95.34069
Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci. 2003; 270(1512):313–21. http://dx.doi.org/10.1098/rspb.2002.2218
Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. Plos Biol. 2004; 2(10):312. http://dx.doi.org/10.1371/journal.pbio.0020312
Herrera-Collazos EE, Galindo-Cuervo AM, Maldonado-Ocampo JA, Rincón-Sandoval M. Three new species of the Eigenmannia trilineata species group (Gymnotiformes: Sternopygidae) from northwestern South America. Neotrop Ichthyol. 2020; 18(1):e180085. http://dx.doi.org/10.1590/10.1590/1982-0224-2018-0085
Hey J. On the failure of modern species concepts. Trends Ecol Evol. 2006; 21(8):447–50. http://dx.doi.org/10.1016/j.tree.2006.05.011
Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001; 17(8):754–55. https://doi.org/10.1093/bioinformatics/17.8.754
Ilves KL, Torti D, López-Fernández H. Exon-based phylogenomics strengthens the phylogeny of Neotropical cichlids and identifies remaining conflicting clades (Cichliformes: Cichlidae: Cichlinae). Mol Phylogenet Evol. 2018; 118:232–43. https://doi.org/10.1016/j.ympev.2017.10.008
Isbrücker IJH. Classification and catalogue of the mailed Loricariidae (Pisces, Siluriformes). Verslagen en Technische Gegevens. Universiteit van Amsterdam. 1980; 22:1–181.
Jarman SN, Elliott NG. DNA evidence for morphological and cryptic Cenozoic speciations in the Anaspididae, “living fossils” from the Triassic. J Evol Biol. 2000; 13(4):624–33. https://doi.org/10.1046/j.1420-9101.2000.00207.x
Jerep FC, Shibatta OA, Pereira EHL, Oyakawa OT. Two new species of Isbrueckerichthys Derijst, 1996 (Siluriformes: Loricariidae) from the rio Paranapanema basin, Brazil. Zootaxa. 2006; 1372(1):53–68. https://doi.org/10.11646/zootaxa.1372.1.5
Kapli P, Lutteropp S, Zhang J, Kobert K, Pavlidis P, Stamatakis A, Flouri T. Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics. 2017; 33(11):1630–38. http://dx.doi.org/10.1093/bioinformatics/btx025
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012; 28(12):1647–49. https://doi.org/10.1093/bioinformatics/bts199
Knowlton N. Sibling species in the sea. Annu Rev Ecol Syst. 1993; 24:189–216. https://doi.org/10.1146/annurev.es.24.110193.001201
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018; 35(6):1547–49. https://doi.org/10.1093/molbev/msy096
Langeani F. Revisão do gênero Neoplecostomus Eigenmann & Eigenmann, 1888, com a descrição de quatro novas espécies do sudeste brasileiro (Ostariophysi, Siluriformes, Loricariidae). Comun Mus Ciênc PUCRS, Sér Zool. 1990; 3(1):3–31.
Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol Evol. 2015; 6(9):1110–16. https://doi.org/10.1111/2041-210X.12410
Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009; 25(11):1451–52. https://doi.org/10.1093/bioinformatics/btp187
López-Fernández H, Honeycutt RL, Winemiller KO. Molecular phylogeny and evidence for an adaptive radiation of geophagine cichlids from South America (Perciformes: Labroidei). Mol Phylogenet Evol. 2005; 34(1):227–44. http://dx.doi.org/10.1016/j.ympev.2004.09.004
López-Fernández H, Winemiller KO, Honeycutt RL. Multilocus phylogeny and rapid radiations in Neotropical cichlid fishes (Perciformes: Cichlidae: Cichlinae). Mol Phylogenet Evol. 2010; 55(3):1070–86. http://dx.doi.org/10.1016/j.ympev.2010.02.020
Lujan NK, Armbruster JW, Lovejoy NR, López-Fernández H. Multilocus molecular phylogeny of the suckermouth armored catfishes (Siluriformes: Loricariidae) with a focus on subfamily Hypostominae. Mol Phylogenet Evol. 2015; 82:269–88. http://dx.doi.org/10.1016/j.ympev.2014.08.020
Luo A, Ling C, Ho SYW, Zhu CD. Comparison of methods for molecular species delimitation across a range of speciation scenarios. Syst Biol. 2018; 67(5):830–46. http://dx.doi.org/10.1093/sysbio/syy011
Lustosa-Costa SY, Ramos TPA, Zawadzki CH, Lima SMQ. Review of the armoured catfish genus Hypostomus (Siluriformes: Loricariidae) from the Parnaíba River basin, Northeastern Brazil, with description of a new species. Neotrop Ichthyol. 2022;20(1):e210126. https://doi.org/10.1590/1982-0224-2021-0126
Mallet J. Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Philos Trans R Soc Lond B Biol Sci. 2008; 363(1506):2971–86. http://dx.doi.org/10.1098/rstb.2008.0081
Mateussi NTB, Melo BF, Foresti F, Oliveira C. Molecular data reveal multiple lineages in piranhas of the genus Pygocentrus (Teleostei, Characiformes). Genes. 2019; 10(5):371. https://doi.org/10.3390/genes10050371
Melo BF, Ochoa LE, Vari RP, Oliveira C. Cryptic species in the Neotropical fish genus Curimatopsis (Teleostei, Characiformes). Zool Scr. 2016; 45(6):650–58. https://doi.org/10.1111/zsc.12178
Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980; 8(19):4321–26. https://doi.org/10.1093/nar/8.19.4321
do Nascimento VD, Coelho KA, Nogaroto V, Almeida RB, Ziemniczak K, Centofante L, Pavanelli CS, Torres RA, Moreira-Filho O, Vicari MR. Do multiple karyomorphs and population genetics of freshwater darter characines (Apareiodon affinis) indicate chromosomal speciation? Zool Anz. 2018; 272:93–103. https://doi.org/10.1016/j.jcz.2017.12.006
Padial JM, Miralles A, De la Riva I, Vences M. The integrative future of taxonomy. Front Zool. 2010; 7:16. http://dx.doi.org/10.1186/1742-9994-7-16
Pereira EHL, Oyakawa OT. Isbrueckerichthys epakmos, a new species of loricariid catfish from the rio Ribeira de Iguape basin, Brazil (Teleostei: Siluriformes). Neotrop Ichthyol. 2003; 1(1):3–09. https://doi.org/10.1590/S1679-62252003000100001
Pereira EH, Reis RE. Revision of the loricariid genera Hemipsilichthys and Isbrueckerichthys (Teleostei: Siluriformes), with descriptions of five new species of Hemipsilichthys. Ichthyol Explor Freshw.2002; 13(2):97–146.
Pereira EHL, Reis RE. Morphology-based phylogeny of the suckermouth armored catfishes, with emphasis on the Neoplecostominae (Teleostei: Siluriformes). Zootaxa. 2017; 4264(1):1–104. http://dx.doi.org/10.11646/zootaxa.4264.1.1
Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol. 2006; 55(4):595–609. https://doi.org/10.1080/10635150600852011
Puillandre N, Brouillet S, Achaz G. ASAP: assemble species by automatic partitioning. Mol Ecol Res. 2020; 21(2):609–20. https://doi.org/10.1111/1755-0998.13281
Puillandre N, Lambert A, Brouillet S, Achaz G. ABGD, automatic barcode gap discovery for primary species delimitation. Mol Ecol. 2012; 21(8):1864–77. https://doi.org/10.1111/j.1365-294X.2011.05239.x
de Queiroz K. Species concepts and species delimitation. Syst Biol. 2007; 56(6):879–86. https://doi.org/10.1080/10635150701701083
Reis RE, Pereira EHL, Armbruster JW. Delturinae, a new loricariid catfish subfamily (Teleostei, Siluriformes), with revisions of Delturus and Hemipsilichthys. Zool J Linn Soc. 2006; 147(2):277–99. https://doi.org/10.1111/j.1096-3642.2006.00229.x
Ribeiro AC. Tectonic history and the biogeography of the freshwater fishes from the coastal drainages of eastern Brazil: an example of faunal evolution associated with a divergent continental margin. Neotrop Ichthyol. 2006; 4(2):225–46. https://doi.org/10.1590/S1679-62252006000200009
Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003; 19(12):1572–74. https://doi.org/10.1093/bioinformatics/btg180
Roxo FF, Albert JS, Silva GSC, Zawadzki CH, Foresti F, Oliveira C. Molecular phylogeny and biogeographic history of the armored Neotropical catfish subfamilies Hypoptopomatinae, Neoplecostominae and Otothyrinae (Siluriformes: Loricariidae). PLoS ONE. 2014; 9(8):105564. https://doi.org/10.1371/journal.pone.0105564
Roxo FF, Ochoa LE, Sabaj MH, Lujan NK, Covain R, Silva GSC, Melo BF, Albert JS, Chang J, Foresti F, Alfaro ME, Oliveira C. Phylogenomic reappraisal of the Neotropical catfish family Loricariidae (Teleostei: Siluriformes) using ultraconserved elements. Mol Phylogenet Evol.2019; 135:148–65. http://dx.doi.org/10.1016/j.ympev.2019.02.017
Roxo FF, Zawadzki CH, Alexandrou MA, Silva GJC, Chiachio MC, Foresti F, Oliveira C. Evolutionary and biogeographic history of the subfamily Neoplecostominae (Siluriformes: Loricariidae). Ecol Evol. 2012; 2(10):2438–49. https://doi.org/10.1002/ece3.368
Roxo FF, Lujan NK, Tagliacollo VA, Waltz BT, Silva GSC, Oliveira C, Albert JS. Shift from slow- to fast-water habitats accelerates lineage and phenotype evolution in a clade of Neotropical suckermouth catfishes (Loricariidae: Hypoptopomatinae). PLoS ONE. 2017; 12(6):0178240. https://doi.org/10.1371/journal.pone.0178240
Santos EO, Deon GA, Almeida RB, Oliveira EA, Nogaroto V, Silva HP, Pavanelli CS, Cestari MM, Bertollo LAC, Moreira-Filho O, Vicari MR. Cytogenetics and DNA barcode reveal an undescribed Apareiodon species (Characiformes: Parodontidae). Genet Mol Biol. 2019; 42(2):365–73. https://doi.org/10.1590/1678-4685-GMB-2018-0066
Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, Crozier RH. Integrative taxonomy: a multisource approach to exploring biodiversity. Annu Rev Entomol. 2010; 55:421–38. https://doi.org/10.1146/annurev-ento-112408-085432
Serrano EA, Melo BF, Freitas-Souza D, Oliveira MLM, Utsunomia R, Oliveira C, Foresti F. Species delimitation in Neotropical fishes of the genus Characidium (Teleostei, Characiformes). Zool Scr. 2018; 48(1):69–80. http://dx.doi.org/10.1111/zsc.12318
Silva-Santos R, Machado CB, Zanata AM, Camelier P, Galetti Jr. PM, Freitas PD. Molecular characterization of Astyanax species (Characiformes: Characidae) from the upper Paraguaçu river basin, a hydrographic system with high endemism. Neotrop Ichthyol. 2023; 21(2):230032. http://dx.doi.org/10.1590/1982-0224-2023-0032
Souza CS, Silva GS, Ochoa LE, Roxo FF, Costa-Silva GJ, Foresti F, Melo BF, Oliveira C. Molecular and morphological diversity in species of Kronichthys (Teleostei, Loricariidae) from Atlantic coastal rivers of Brazil. J Fish Biol. 2021; 98(3):668–79. https://doi.org/10.1111/jfb.14607
Souza TB, Ferreira DC, Silva HP, Netto-Ferreira AL, Venere PC. DNA Barcoding of Pyrrhulina australis (Characiformes: Lebiasinidae) reveals unexpected cryptic diversity in the group. Neotrop Ichthyol. 2023; 21(4):e230037. https://doi.org/10.1590/1982-0224-2023-0037
de Sousa JLP, Bitencourt JA, Sampaio I, Schneider H, Affonso PRAM. “More than meets the eye”: phylogeographic inferences and remarkable cryptic diversity and in endemic catfish Parotocinclus (Loricariidae: Hypoptopomatinae) from neglected and impacted basins in South America. Conserv Genet. 2021; 22:411–25. https://doi.org/10.1007/s10592-021-01336-3
Tang CQ, Humphreys AM, Fontaneto D, Barraclough TG. Effects of phylogenetic reconstruction method on the robustness of species delimitation using single-locus data. Methods Ecol Evol. 2014; 5(10):1086–94. http://dx.doi.org/10.1111/2041-210x.12246
Traldi JB, Vicari MR, Martinez JF, Blanco DR, Lui RL, Azambuja M, Almeida RB, Malimpensa GC, Costa Silva GJ, Oliveira C, Pavanelli CS, Moreira-Filho O. Apareiodon species evolutionary divergence (Characiformes: Parodontidae) evidenced by chromosomal and molecular inference. Zool Anz. 2020; 289:166–76. http://dx.doi.org/10.1016/j.jcz.2020.10.010
Ward RD. DNA barcode divergence among species and genera of birds and fishes. Mol Ecol Res. 2009; 9(4):1077–85. http://dx.doi.org/10.1111/j.1755-0998.2009.02541.x
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia’s fish species. Philos Trans R Soc Lond B Biol Sci. 2005; 360(1462):1847–57. https://doi.org/10.1098/rstb.2005.1716
Wiley EO. The evolutionary species concept reconsidered. Syst Zool. 1978; 27(1):17–26. http://dx.doi.org/10.2307/2412809
Willis SC. One species or four? Yes!…and, no. Or, arbitrary assignment of lineages to species obscures the diversification processes of Neotropical fishes. PLoS ONE. 2017; 12(2):e0172349. http://dx.doi.org/10.1371/journal.pone.0172349
Zapata F, Jiménez I. Species delimitation: inferring gaps in morphology and geography. Syst Biol. 2011; 61(2):179–94. http://dx.doi.org/10.1093/sysbio/syr084
Zhang J, Kapli P, Pavlidis P, Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013; 29(22):2869–76. https://doi.org/10.1093/bioinformatics/btt499
Authors
![]() Rafael B. de Almeida1,
Rafael B. de Almeida1, ![]() Matheus Azambuja1,
Matheus Azambuja1, ![]() Viviane Nogaroto2,
Viviane Nogaroto2, ![]() Claudio Oliveira3,
Claudio Oliveira3, ![]() Fábio F. Roxo3,
Fábio F. Roxo3, ![]() Cláudio H. Zawadzki4 and
Cláudio H. Zawadzki4 and ![]() Marcelo R. Vicari1,2
Marcelo R. Vicari1,2 ![]()
[1] Programa de Pós-Graduação em Genética, Universidade Federal do Paraná, Centro Politécnico, Jardim das Américas, 81531-990 Curitiba, PR, Brazil. (RBA) bonfimetal@hotmail.com, (MA) matheus_azambuja@hotmail.com, (MRV) vicarimr@uepg.br (corresponding author).
[2] Departamento de Biologia Estrutural, Molecular e Genética, Universidade Estadual de Ponta Grossa, Av. Carlos Cavalcanti, 4748, 84030-900 Ponta Grossa, PR, Brazil. (VN) vivianenogaroto@hotmail.com.
[3] Laboratório de Biologia e Genética de Peixes, Departamento de Biologia Estrutural e Funcional, Instituto de Biociências de Botucatu, Universidade Estadual Paulista, R. Prof. Dr. Antônio C. W. Zanin, s/n, Rubião Jr., 18618-689 Botucatu, SP, Brazil. (CO) claudio.oliveira@unesp.br, (FFR) roxoff@hotmail.com.br.
[4] Núcleo de Pesquisas em Limnologia, Ictiologia e Aquicultura (Nupélia), Departamento de Biologia, Universidade Estadual de Maringá, Av. Colombo, 5790, 87020-900 Maringá, PR, Brazil. (CHZ) chzawadzki@hotmail.com.
Authors’ Contribution 

Rafael B. de Almeida: Formal analysis, Investigation, Methodology, Writing-original draft.
Matheus Azambuja: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing-original draft, Writing-review and editing.
Viviane Nogaroto: Data curation, Formal analysis, Investigation, Methodology, Supervision, Visualization, Writing-original draft, Writing-review and editing.
Claudio Oliveira: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Validation, Visualization, Writing-original draft, Writing-review and editing.
Fábio F. Roxo: Data curation, Formal analysis, Investigation, Methodology.
Cláudio H. Zawadzki: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing-original draft, Writing-review and editing.
Marcelo R. Vicari: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing-original draft, Writing-review and editing.
Ethical Statement
Fishes were collected with authorization of the Instituto Chico Mendes de Conservação da Biodiversidade (ICMBIO), Sistema de Autorização e Informação em Biodiversidade (SISBIO – License Ids 13843–6 and 15117–4). This study procedures agree with the Ethics Committee of Animal Usage of the Universidade Estadual de Ponta Grossa (Protocol: 06/2019).
Competing Interests
The author declares no competing interests.
How to cite this article
Almeida RB, Azambuja M, Nogaroto V, Oliveira C, Roxo FF, Zawadzki CH, Vicari MR. DNA barcode shows discordant cases among morphological and molecular species identification in Isbrueckerichthys (Siluriformes: Loricariidae). Neotrop Ichthyol. 2024; 22(3):e240040. https://doi.org/10.1590/1982-0224-2024-0040
Copyright
This is an open access article under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in any medium, provided the original work is properly cited.
Distributed under
Creative Commons CC-BY 4.0

© 2024 The Authors.
Diversity and Distributions Published by SBI
![]() Accepted August 5, 2024 by Hernán López-Fernández
Accepted August 5, 2024 by Hernán López-Fernández
![]() Submitted May 7, 2024
Submitted May 7, 2024
![]() Epub October 18, 2024
Epub October 18, 2024